Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming (original) (raw)
. Author manuscript; available in PMC: 2016 Dec 3.
Published in final edited form as: Cell Stem Cell. 2015 Oct 29;17(6):689–704. doi: 10.1016/j.stem.2015.09.005
SUMMARY
Epigenetic and epitranscriptomic networks have important functions in maintaining pluripotency of embryonic stem cells (ESCs) and somatic cell reprogramming. However the mechanisms integrating the actions of these distinct networks are only partially understood. Here, we show that the chromatin-associated zinc finger protein 217 (ZFP217) coordinates epigenetic and epitranscriptomic regulation. ZFP217 interacts with several epigenetic regulators, activates transcription of key pluripotency genes, and modulates N6-methyladenosine (m6A) deposition on their transcripts by sequestering the enzyme m6A methyltransferase-like 3 (METTL3). Consistently, Zfp217 depletion compromises ESC self-renewal and somatic cell reprogramming, globally increases m6A RNA levels, and enhances m6A modification of Nanog, Sox2, Klf4, and c-Myc mRNAs, promoting their degradation. ZFP217 binds its own target gene mRNAs, which are also METTL3-associated, and is enriched at promoters of m6A-modified transcripts. Collectively, these findings shed light on how a transcription factor can tightly couple gene transcription to m6A RNA modification to insure ESC identity.
INTRODUCTION
Embryonic stem cell (ESC) self-renewal and somatic cell reprogramming require precise coordination of transcription factors, chromatin regulators and RNA modifiers to be sustained. Considerable efforts have been devoted to characterize the epigenetic and transcription networks controlling pluripotency. However, the function of post-transcriptional RNA modifications maintaining the equilibrium between ESC self-renewal and differentiation remains poorly understood.
Zinc finger protein 217 (Zfp217) encodes a transcription factor with eight conserved C2H2 zinc finger motifs and a proline-rich transactivation domain. Overexpression and/or genetic amplification of the human homolog ZNF217 correlates with poor survival in a variety of cancers [reviewed by (Quinlan et al., 2007)]. Human mammary epithelial cells or ovarian cells transduced with ZNF217 bypass senescence and achieve an immortalized state (Li et al., 2007; Nonet et al., 2001), a hallmark of both cancer and stem cells. Overexpression of ZNF217 provides a selective advantage to tumor cells by deregulating pathways associated with normal growth, apoptosis or differentiation (Huang et al., 2005; Thollet et al., 2010), in part through INK4B locus repression (Thillainadesan et al., 2012). ZNF217 has also been associated with Aurora kinase A overexpression (Thollet et al., 2010) and with the activation of the TGF-β (Vendrell et al., 2012) and the AKT pathways (Huang et al., 2005), required to maintain pluripotency (Boiani and Scholer, 2005; Lee et al., 2012; Welham et al., 2011). Moreover, induced differentiation of Ntera2 cells with retinoic acid leads to downregulation of ZNF217 (Krig et al., 2007). Despite increasing observations supporting a function of ZNF217 in the maintenance of the undifferentiated state, the role of this transcription factor in ESC biology remains unexplored.
Recent studies propose that _N_6-methyladenosine (m6A), the most abundant post-transcriptional modification in RNA (Jia et al., 2013; Tuck, 1992), can determine the fate of ESC self-renewal and pluripotency (Wang et al., 2014b), whereas others postulate that m6A is required for transition to differentiated states (Batista et al., 2014; Geula et al., 2015). The core mammalian methyltransferase complex includes the methyltransferase-like 3 (METTL3, also known as MT-A70) and the methyltransferase-like 14 (METTL14), which associate with additional regulatory factors such as Wilm's tumor 1 associating protein (WTAP) (Liu et al., 2014; Ping et al., 2014). Enrichment of m6A in RNA effectively influences all aspects of RNA metabolism, including mRNA stability (Wang et al., 2014a; Wang et al., 2014b), alternative splicing (Fustin et al., 2013; Geula et al., 2015; Ping et al., 2014; Zhao et al., 2014), mRNA translation efficiency (Geula et al., 2015), and secondary RNA structure and localization, resulting in alterations in an array of cellular processes. Nonetheless, the molecular function of m6A RNA modification in development remains incompletely understood.
In the present study we elucidate the multi-faceted role of ZFP217 in pluripotency. We show that ZFP217 directly activates the transcription of Nanog, Sox2, and many other genes linked to the undifferentiated state. Furthermore, ZFP217 restricts the deposition of _N_6-methyladenosine (m6A) at these transcripts through METTL3 interaction. Depletion of Zfp217 results in a decrease in the expression of pluripotency factors and a global increase of m6A methylation, promoting degradation of core stem cell transcripts. Taken together, these findings demonstrate that ZFP217 is an essential regulator that balances self-renewal and differentiation not just by regulating the epigenome but also the epitranscriptome of pluripotency-associated factors.
RESULTS
Loss of Zfp217 impairs self-renewal and triggers differentiation in ESCs
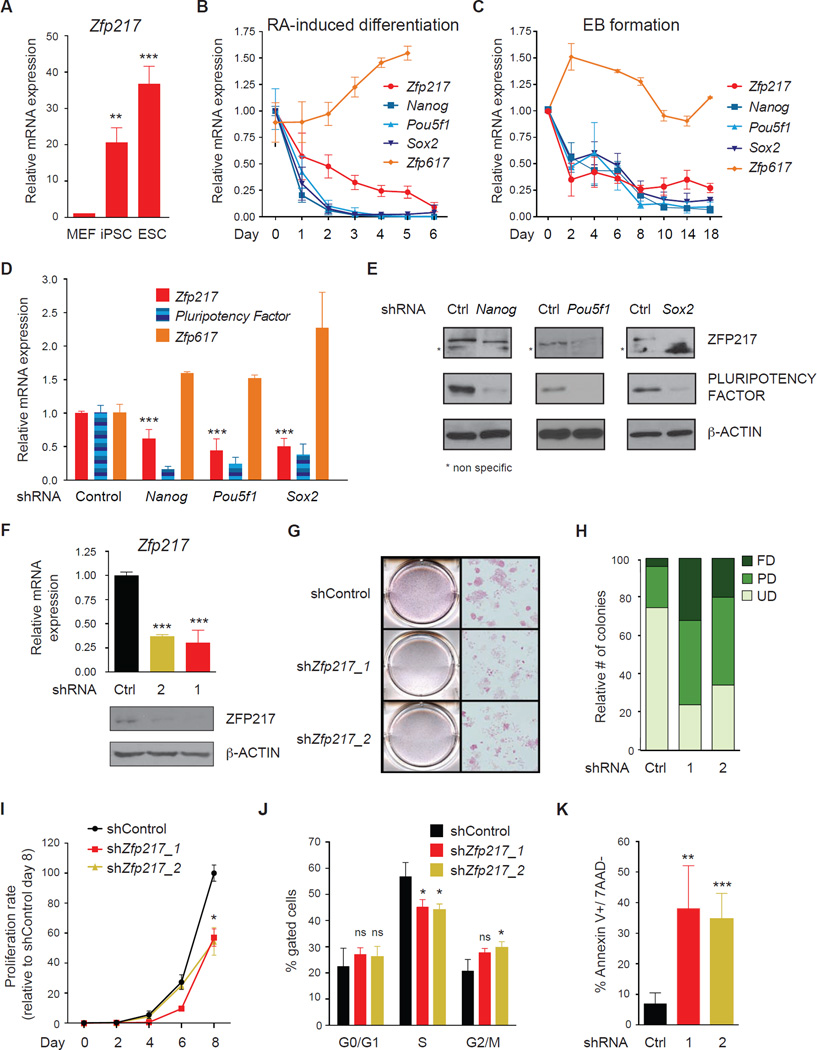
To explore the function of ZFP217 in ESCs, we analyzed the expression of Zfp217 in mouse embryonic fibroblasts (MEFs), ESCs, and induced pluripotent stem cells (iPSCs) by quantitative PCR with reverse transcription (RT–qPCR). Zfp217 was significantly enriched in ESCs and iPSCs compared to MEFs (Figure 1A). Examination of Zfp217 expression in retinoic acid (RA)-induced differentiation and in embyoid bodies (EBs) revealed a gradual decrease in Zfp217 levels, but not Zfp617, along the course of differentiation, that correlated with the decrease of Nanog, Pou5f1 (also known as Oct3/4), and Sox2 (Figures 1B and 1C). In agreement with these findings, Zfp217 RNA and ZFP217 protein levels were decreased upon short hairpin RNA (shRNAs) of the pluripotency factors Nanog, Pou5f1 and Sox2 (Figures 1D and 1E). Next, we conducted loss-of-function assays by using two distinct shRNAs that exhibited at least 80% knockdown of endogenous ZFP217 protein and Zfp217 RNA in ESCs (Figure 1F). While control cells retained ESC morphology, _Zfp217_-depleted cells exhibited a flattened fibroblastic-like morphology typical of differentiating ESCs, with decreased levels of alkaline phosphatase (AP) staining (Figure 1G). Specifically, Zfp217 depletion increased the number of partially and fully differentiated colonies, whereas the number of undifferentiated colonies drastically decreased (Figure 1H). Furthermore, loss of Zfp217 resulted in severely compromised cell growth (Figure 1I), impaired cell cycle profile (Figure 1J) and marked increase in early apoptosis (Figure 1K). Taken together, these results suggest that ZFP217 is required to maintain the pluripotency state of ESCs.
Figure 1. ZFP217 is required to maintain the pluripotent state of ESCs.
(A) RT-qPCR analysis of Zfp217 expression in MEFs, iPSCs and ESCs. Data are represented as mean ± SD; n = 3. ***p<0.0005; **p<0.005 versus MEFs.
(B and C) RT-qPCR analysis of Zfp217, Pou5f1, Sox2, and Zfp617 during RA-induced differentiation (B) and EB formation (C). Data are represented as mean ± SD; n = 3.
(D–E) RT-qPCR analysis of Zfp217 (D) and representative western blot (E) for ZFP217 upon Nanog, Pou5f1 and Sox2 depletion. The expression of the pluripotency factors was determined to monitor the knockdown efficiency. Error bars show ± SD; n = 3. ***p<0.0001 versus control shRNA. For the immunoblots β-ACTIN was used as a loading control. * non specific.
(F) RT-qPCR (upper panel) and western blot analysis (lower panel) to monitor Zfp217 knockdown efficiency. Error bars show ± SD; n = 3. ***p<0.0001 versus control shRNA. For the immunoblot, β-ACTIN was used as a loading control.
(G–H) AP staining of control and _Zfp217_-depleted ESCs (G). Percentage of ESC colonies were counted and depicted in the graph (H). Undifferentiated (UD); partially differentiated (PD); fully differentiated (FD).
(I) Proliferation rate of control and _Zfp217_-depleted ESCs relative to shRNA control at day 8. Error bars show ± SD; n = 3. *p=0.01 versus control shRNA.
(J) Cell cycle distribution of control and _Zfp217_-depleted ESCs examined by DNA content index. Error bars show ± SD; n = 3. ns= not significant; *p<0.05 versus control.
(K) Percentage of early apoptotic cells in control and _Zfp217_-depleted ESCs defined as Annexin V-positive and 7AAD-negative cells. Error bars show ± SD; n = 3. **p<0.005; ***p<0.001 versus control shRNA.
Somatic cell reprogramming is impaired upon Zfp217 knockdown
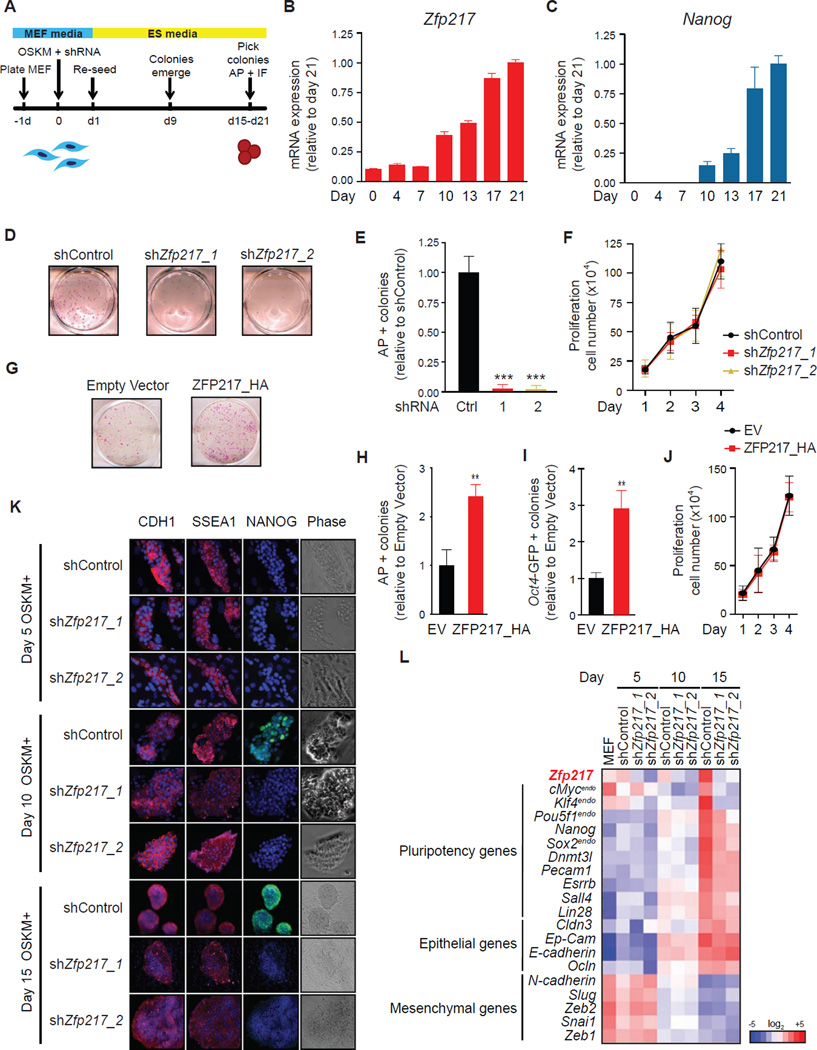
Given the role of ZFP217 in the maintenance of ESC identity, we examined ZFP217 function during iPSC reprogramming. We transduced MEFs by lentiviral infection with a polycistronic cassette constitutively expressing the Yamanaka factors (OCT4, SOX2, KLF4 and c-MYC; OSKM) (Figure 2A) (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), and we observed upregulation of both Zfp217 and Nanog during mouse iPSC generation (Figures 2B and 2C). Next, the ectopic expression of OSKM was combined with either control or Zfp217 shRNA, being the transduction efficiency comparable in both conditions (Figure S1A). Depletion of Zfp217 in reprogrammable MEFs markedly reduced the number of AP-positive iPSC colonies, suggesting that ZFP217 plays a positive role in iPSC generation (Figures 2D and 2E). Importantly, depletion of Zfp217 did not affect the proliferation of reprogramming MEFs (Figure 2F), indicating that decrease in iPSC number is not due to adverse effects on proliferative capacity of transduced MEFs. Isolated iPSC clones originated from Zfp217 shRNA transduced MEFs were not bona fide iPSCs, as they exhibited a significantly decreased expression of Zfp217 and Nanog relative to control shRNA (Figure s S1B and S1C). A positive role in reprogramming was further supported by ZFP217 overexpression with substantial increase in iPSC colony numbers (Figures 2G–J). Reprogramming is a multi-step process, initiated by enhanced proliferation, followed by a mesenchymal-to-epithelial transition (MET) (Li et al., 2010), and completed by activation of pluripotency genes. In order to identify what stage of reprogramming was blocked upon Zfp217 depletion, we performed immunofluorescence microscopy for E-CADHERIN (CDH1), the hallmark of epithelial cell identity, the stem-cell marker stage-specific embryonic antigen 1 (SSEA1), and NANOG at days 5, 10, and 15 after OSKM reprogramming in the presence of control or Zfp217 shRNA. CDH1 and SSEA1 were activated by day 5 post-infection even in the absence of ZFP217 (Figure 2K). Conversely, NANOG was not activated at day 10 post-transduction, and its expression remained undetectable at day 15 in reprogramming MEFs depleted of Zfp217 (Figure 2K). Consistently, the elevation of several pluripotency genes was impaired upon loss of Zfp217 whereas the expression of most epithelial and mesenchymal genes analyzed was unaffected in _Zfp217_-depleted reprogramming MEFs compared to control (Figures 2L and S1D–O). Overall, our results indicate that ZFP217 is required for generation of iPSCs from MEFs, being predominantly indispensable for later stages of somatic cell reprogramming, when activation of the pluripotency factors occurs.
Figure 2. ZFP217 is required for somatic cell reprogramming.
(A) Schematics of iPSC generation.
(B–C) Representative RT-qPCR analysis for Zfp217 (B) and Nanog (C) during MEF reprogramming.
(D–F) AP staining (D) and quantification (E) of control and _Zfp217_-depleted iPSCs at day 21. Error bars show ± SD; n = 3. ***p<0.0001 versus control shRNA. (F) Proliferation rate of control and _Zfp217_-depleted reprogramming MEFs.
(G–J) AP staining of reprogramming MEFs overexpressing ZFP217_HA or empty vector at day 15 post-transduction (G). Number of AP+ (H) and GFP+ (I) colonies in ZFP217_HA relative to empty vector. Error bars show ± SD; n = 3. **p<0.001 versus empty vector. (J) Proliferation rate of reprogramming MEFs transduced with ZFP217_HA or empty vector.
(K) E-CADHERIN (CDH1), SSEA1 and NANOG immunostaining at indicated days of reprogramming with control or Zfp217 shRNA. Bright field is depicted in the right panel. The scale bar represents 100 µm.
(L) Heat map illustrating the relative expression of pluripotency, epithelial and mesenquimal genes measured by RT-qPCR in reprogramming MEFs with control or Zfp217 shRNA at indicated days.
ZFP217 positively regulates the ESC transcriptome
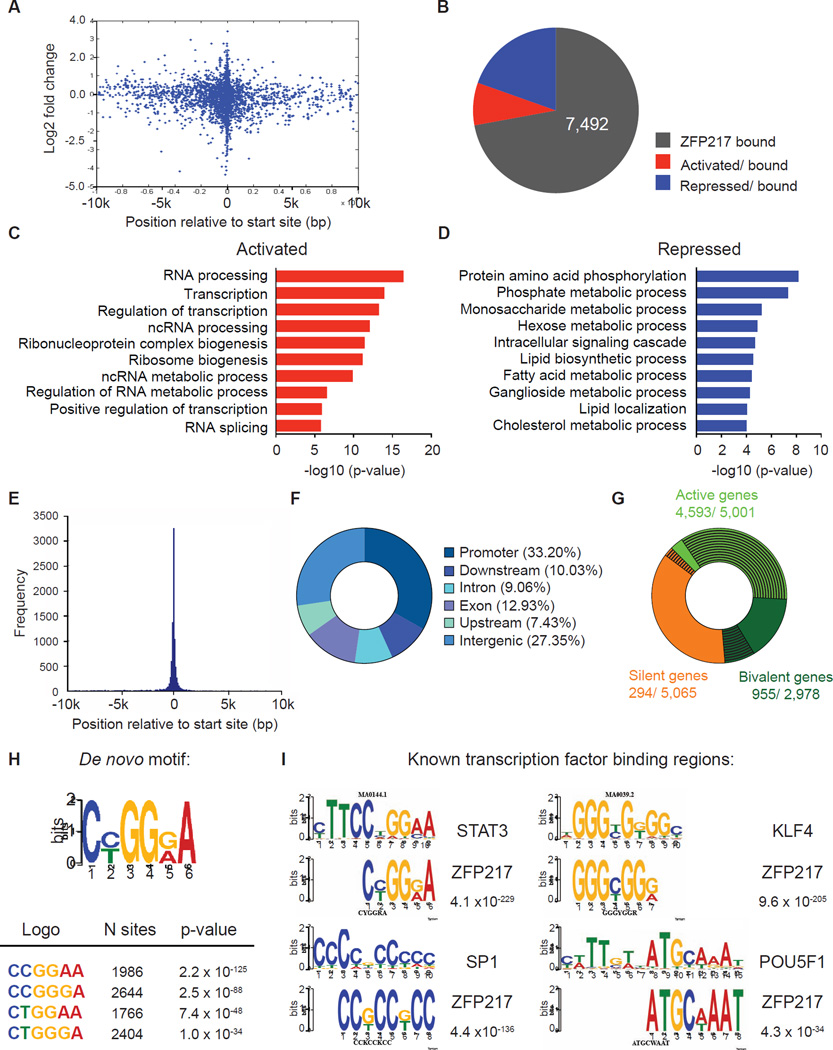
To gain an overview of the global role of ZFP217 in pluripotency, we performed chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) and RNA sequencing (RNA-Seq) in control and Zfp217 knockdown ESCs. Analysis of ZFP217 binding identified 10,406 target genes (Table S1) that were more likely to be regulated upon Zfp217 knockdown if the binding peak was located at the transcription start site (TSS) (Figure 3A). Nevertheless, despite all regulated genes in _Zfp217_-depleted ESCs were bound by ZFP217 they represented only a minority of ZFP217 ChIP-Seq targets (Figure 3B), and were not associated with genes important for ESC identity (data not shown). RNA-Seq analysis revealed 882 upregulated genes (Table S1) in _Zfp217_-depleted ESCs that associated with gene ontology (GO) categories related to RNA metabolic processes and transcription (Figure 3C), whereas the 2,058 downregulated genes upon Zfp217 knockdown (Table S1) were involved in diverse functions (Figure 3D). Analysis of peak distribution showed that ZFP217 predominantly occupied promoters (within 5 Kb of annotated TSS) and intergenic regions of actively transcribed and bivalent genes (Figures 3E–G and S2A–C). Because human ZNF217 is associated with the corepressor for element-1-silencing transcription factor (CoREST) (Cowger et al., 2007; Lee et al., 2005; You et al., 2001), we examined the overlap between ZFP217 and the previously published REST cofactors (SIN3A/B) and CoREST complex (RCOR1, RCOR2, RCOR3) datasets. Our analysis revealed that the vast majority of genes occupied by some of the CoREST members were enriched with ZFP217 (Figure S2D). Accordingly with the fact that the CoREST complex is implicated in neuronal differentiation (Yu et al., 2011), the ZFP217-bound bivalent genes functioned in commitment to the neural lineage (Figure S2E).
Figure 3. ZFP217 is associated with both promoters and enhancers in ESC.
(A) Graph indicating the relative position of the unique closest ZFP217 peaks with respect to the TSS (x-axis) and the log2 fold change in gene expression in response to Zfp217 depletion (y-axis).
(B) Pie chart depicting the number of ZFP217 only bound genes, activated and bound, and repressed and bound in _Zfp217_-depleted ESCs.
(C–D) Functional categories of genes activated and bound (C) and repressed and bound (D) upon Zfp217 depletion of ESCs showing the p-value for the enrichment of biological process GO-term.
(E) Histogram showing the distribution of ZFP217-binding peaks relative to the nearest TSS.
(F) Pie chart of the genomic distribution of ZFP217-binding peaks including promoters (within 5 kb upstream of TSS), downstream (within 10 kb downstream of the gene), introns, exons, upstream (within 10 kb upstream of the gene), and intergenic regions.
(G) Pie chart depicting ZFP217 binding sites (black lines) at active (green), bivalent (dark green) and silent (orange) genes (defined in Supplemental Experimental Procedures).
(H–I) ZFP217 de novo motif (H) and ZFP217 binding at known binding motifs for STAT3, KLF4, SP1 and POU5F1 (I) with the corresponding p-values.
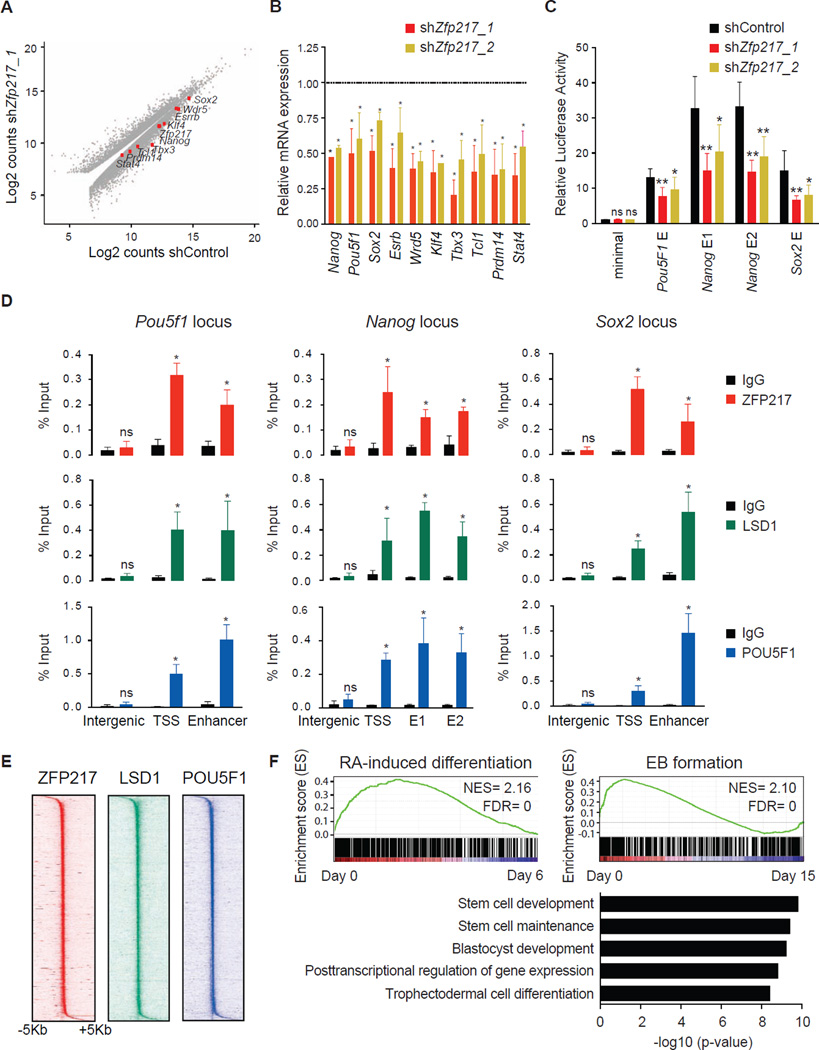
Next, we performed de novo motif analysis using the algorithm MEME, and identified a unique DNA motif enriched at ZFP217 sites (Figure 3H), which overlapped with the binding sequence for STAT3 (Figure 3I). Secondary motifs overrepresented in ZFP217-bound regions included known binding sequences for KLF4, SP1 and POU5F1 (Figure 3I), among others, suggesting that ZFP217 might occupy an overlapping set of genomic sites important for the regulation of the undifferentiated state. In agreement with this observation, inspection of individual gene tracks of ZFP217 and public available POU5F1 datasets showed ZFP217 and POU5F1 binding at the core promoter and at the enhancers of Pou5f1, Nanog and Sox2 (Figure s S3A–C). Moreover, RNA-Seq profiling revealed a significant decrease of key ESC transcripts upon Zfp217 depletion (Figures 4A and 4B), suggesting an important role of ZFP217 in regulating the expression of stem cell identity genes. Further expression analysis revealed that levels of ectodermal and endodermal markers, but not trophectodermal and mesodermal markers, were increased in Zfp217 knockdown ESCs (Figure s S3D–G), suggesting that the repressor activity of ZFP217 on developmental genes is also important for ESC maintenance. To further determine whether ZFP217 directly regulates the expression of genes encoding the core pluripotency factors, we cloned the well characterized Nanog, Pou5f1 and Sox2 enhancers into Oct4 minimal promoter, and transfected these reporters into either control or _Zfp217_-depleted ESCs. A significant decrease in luciferase activity upon Zfp217 knockdown suggested a direct regulation (Figure 4C), which was further validated by ChIP-qPCR of ZFP217 (Figure 4D). Because lysine demethylase 1 (LSD1), a known interacting partner of ZFP217 involved in enhancer decommissioning of pluripotency genes (Whyte et al., 2012), also bound these genomic regions (Figure 4D and S3A–C), we reasoned that the overlap of ZFP217, LSD1 and POU5F1 genome-wide binding might reveal the occupancy of the stem cell identity genes. Indeed, ZFP217, LSD1 and POU5F1 co-occupied hundreds of sites in the ESC genome (Figure S3H), which was supported by heat map clustering of the read density, where an enriched signal was prevalent surrounding the center of ZFP217 binding sites (Figure 4E). Nevertheless, additional elements must be required for this association as co-immunoprecipitation experiments showed interaction between ZFP217 and LSD1, but not with POU5F1 (Figure S3I). Expectedly, ZFP217-LSD1-POU5F1 target genes were enriched in undifferentiated cells compared to RA-induced differentiated cells (Table S2) or EBs (Figure 4F, upper panel), and were associated with an ESC-identity signature (Figure 4F, lower panel). Accordingly, ZFP217 localized at super enhancers, identified at genes important for the maintenance of pluripotency (Figure S3J) (Whyte et al., 2013). Overall, our findings indicate that ZFP217 globally contributes to the transcriptional program that maintains the expression of core stem cell genes to preserve ESC self-renewal.
Figure 4. ZFP217 positively regulates the ESC transcriptome.
(A) Scatter plot of upregulated and downregulated genes in control compared to _Zfp217_-depleted ESCs.
(B) RT-qPCR analysis of pluripotency-associated genes in ESCs transduced with Zfp217_1 or Zfp217_2 compared to control shRNA. Error bars show ± SD; n = 3. *p<0.0001 versus control shRNA.
(C) Luciferase assay of the indicated constructs transfected in control or _Zfp217_-depleted ESCs. Data is normalized to CMV-renilla luciferase and represented relative to the minimal Oct4 promoter. Error bars show ± SD; n = 3. **p<0.001; *p<0.05; ns = not significant versus control shRNA.
(D) ChIP-qPCR analysis of ZFP217 (upper panel), LSD1 (medium panel) and POU5F1 (lower panel) binding at the Pou5f1, Nanog and Sox2 locus. The positions of the amplified regions are indicated at Figures S3A–C. TSS, transcriptional start site; E1 and E2, enhancer 1 and enhancer 2, respectively. Error bars show ± SD; n = 3. *p<0.0001; ns = not significant versus IgG control.
(E) Density maps of ZFP217, LSD1, and POU5F1 ChIP-Seq datasets at TSS. Color scale indicates ChIP-Seq signal in reads per million.
(F) GSEA plots of ZFP217, LSD1, and POU5F1-bound genes during RA-induced differentiation and EB formation. High and low expression of genes is represented in red and blue, respectively. NES, normalized enrichment score; FDR, false discovery rate (upper panel). Functional categories of ZFP217, LSD1, and POU5F1-bound genes using GREAT software (lower panel).
ZFP217 interacts with METTL3 and negatively affects m6A deposition
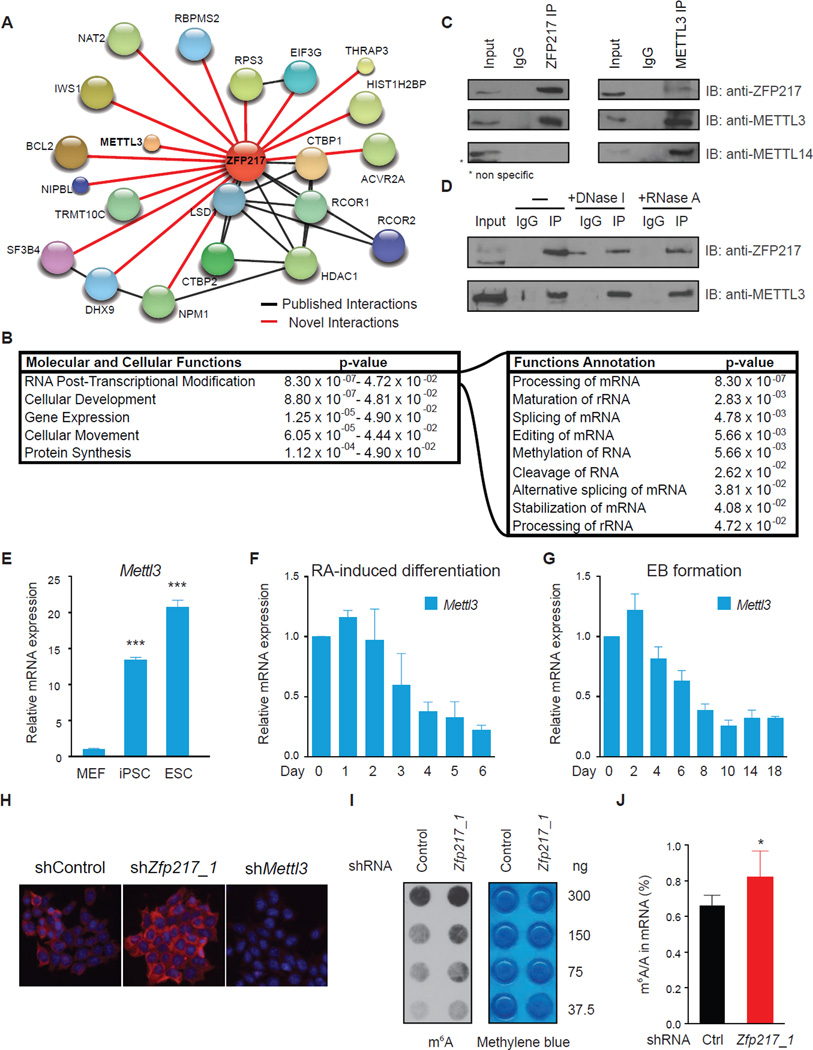
To gain insight into the molecular mechanism by which ZFP217 controls ESC maintenance and somatic cell reprogramming, we performed immunoprecipitation experiments with endogenous ZFP217 in ESCs, followed by liquid chromatography tandem mass spectrometry (LC-MS/MS). We identified ZFP217 as well as peptides representative of LSD1, RCOR1, RCOR2, C-terminal-binding protein 1 (CTBP1) and 2 (CTBP2), and histone deacetylase 1 (HDAC1), consistent with previously reported binding partners of ZNF217 and thus validating our approach (Banck et al., 2009; Cowger et al., 2007; Quinlan et al., 2006; You et al., 2001) (Figures 5A and Table S3). Interestingly, we also identified numerous proteins involved in RNA post-transcriptional modification, including mRNA and rRNA processing, rRNA maturation and splicing, and methylation, among others (Figure 5B).
Figure 5. ZFP217 interacts with METTL3 and counteracts its activity.
(A) Diagram depicting the network of associated proteins identified through LC-MS/MS of ZFP217. Black and red lines represent published and novel interactions, respectively.
(B) IPA analysis of proteins identified with LC-MS/MS. Left box describes the molecular and cellular functions. Right box describes function annotations of the RNA post-transcriptional modification category.
(C) Immunoprecipiation of nuclear extracts from ESCs with antibodies against ZFP217 (left panel) or METTL3 (right panel) followed by immunoblotting with ZFP217, METTL3 and METTL14 antibodies. IgG was used as a control.
(D) Immunoprecipiation of nuclear extracts pretreated with DNase I or RNase A with antibodies against ZFP217 followed by immunoblotting with ZFP217 and METTL3 antibodies. IgG was used as a control.
(E) RT-qPCR analysis of Mettl3 expression in MEFs, iPSCs and ESCs. Error bars show ± SD; n = 3. ***p<0.0005 versus MEFs.
(F and G) RT-qPCR analysis of Mettl3 during RA-induced differentiation (F) and EB formation (G). Error bars show ± SD; n = 3
(H) m6A immunostaining of ESCs transduced with control, Zfp217_1, or Mettl3_1 shRNAs. Nuclei were stained with DAPI. The scale bar represents 100 µm
(I) Dot blot analysis of polyadenylated RNA (poly(A)+) isolated from control and Zfp217_1 shRNA ESCs. Indicated amounts were loaded and detected with m6A antibody. Methylene blue staining was used as a loading control.
(J) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from control and Zfp217_1 knockdown ESCs. Error bars show ± SD; n = 2. *p<0.05 versus control shRNA.
We focused our attention on the interaction of ZFP217 with the RNA methyltransferase METTL3 because the regulation and function of m6A mRNA in ESCs remains largely unknown. Although ZFP217 has not been identified as a METTL3-interacting protein in recent studies (Liu et al., 2014; Schwartz et al., 2014), we could verify a robust and specific association between ZFP217 and METTL3 in ESCs by co-immunoprecipitation (Figure 5C; left panel) and reverse Co-IP experiments of the endogenous proteins (Figure 5C; right panel). Consistent with the fact that METTL14 was not present in our LC-MS/MS analysis, we could not detect METTL14 after ZFP217 immunoprecipitation (Figure 5C; bottom panel), suggesting that METTL3 could be part of multiple dynamic complexes. Notably, ZFP217-METTL3 association was DNA and RNA-independent (Figure 5D), as treatment of the nuclear extracts with DNase I or RNase A did not interfere with the precipitation of METTL3 with anti-ZFP217 antibodies.
Mettl3 expression was markedly enriched in ESCs and iPSCs compared to MEFs (Figure 5E). Accordingly, Mettl3 mRNA and the mRNA of other members of the m6A methyltransferase complex, was significantly decreased upon RA and EB-induced ESC differentiation (Figures 5F, 5G and S4A). However, _Mettl3_-depleted ESCs maintained an undifferentiated state, with high AP activity and no significant changes in the core stem cell factors expression (Figure s S4B–D), suggesting that METTL3 is dispensable for ESC self-renewal and may be involved in other aspects of stem cell biology as previously proposed (Batista et al., 2014). Because Mettl3 RNA and METTL3 protein levels were unchanged upon Zfp217 knockdown (Figure S4E), we sought to analyze whether ZFP217 was affecting METTL3 m6A activity in vivo. Thus, we analyzed m6A methylation levels in control and _Zfp217_-depleted ESCs by using three independent methods: m6A immunostaining, dot blot, and LC-MS/MS quantification of the m6A/A ratios in purified polyadenylated (poly(A)+) RNA. Our analysis revealed a global increase of m6A levels in _Zfp217_-depleted cells compared to control (Figures 5H–J and S4F–H), whereas m6A levels were drastically decreased upon Mettl3 knockdown (Figures 5H and S4I). Taken together, our data suggest that ZFP217 interacts with METTL3 to prevent cellular m6A RNA deposition.
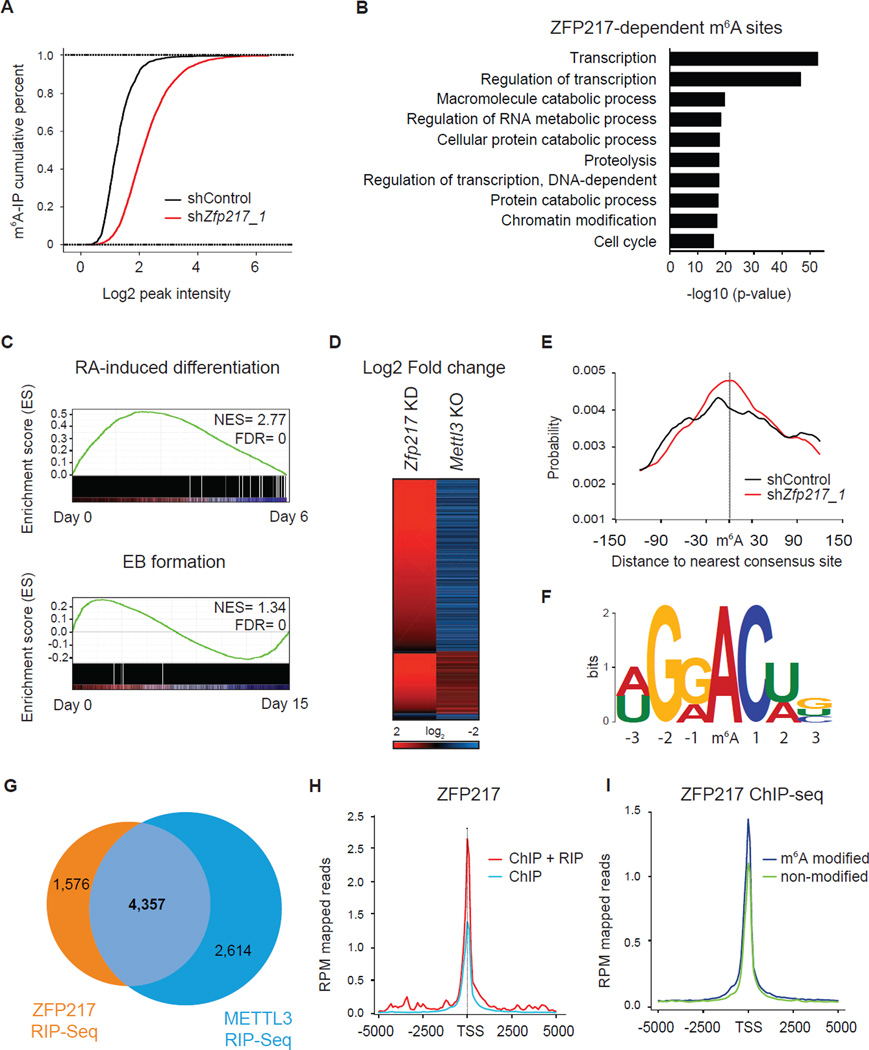
Analysis of m6A methylome in ESCs depleted of Zfp217
To elucidate the mechanism by which ZFP217 regulates m6A restriction, we analyzed m6A RNA methylation in control and _Zfp217_-depleted ESCs through methylated RNA immunoprecipitation sequencing (MeRIP–Seq) (Table S4). Comparison between the two conditions revealed an increase of m6A sites across 3,586 RNAs upon Zfp217 knockdown (Figure 6A). Henceforth, we will refer to these transcripts as the ZFP217-dependent sites. ZFP217-dependent RNAs spanned a wide set of functions (Figure 6B), which were enriched in the undifferentiated state assessed by GSEA analysis (Figure 6C). Next, we overlapped ZFP217-dependent sites with published available MeRIP-Seq datasets of Mettl3 knockout ESCs (Batista et al., 2014). The vast majority of the common peaks (72%) experienced a decrease in m6A methylation levels upon Mettl3 knockout and an increase in m6A RNA modification after _Zfp217_-depletion, suggesting that ZFP217 is antagonizing the RNA methyltransferase activity of METTL3 (Figure 6D). Interestingly, 25% of common peaks underwent increase in m6A levels in Zfp217 and _Mettl3_-depleted cells, whereas just 3% of them had a decrease methylation in both conditions. Methylated m6A sites from control and _Zfp217_-depleted cells were located within a median distance of 30 nt from the nearest consensus RRACU sequence motif (Figure 6E–6F) (Meyer and Jaffrey, 2014). As previously described (Dominissini et al., 2012; Meyer et al., 2012), m6A sites were significantly enriched near the stop codon and the beginning of the 3′ UTR, with virtually no peaks located at the TSS of the protein coding genes (Figure S5A). The m6A enrichment was positively correlated with the exon length, being median internal exons harboring sites in Zfp217 shRNA ESCs longer than in control shRNA (Figure S5B). In order to test if the global increase in m6A levels was a consequence of increased transcription upon Zfp217 knockdown, we analyzed the expression of transcripts harboring m6A modification in control and _Zfp217_-depleted ESCs. Our analysis revealed that these transcripts were not differentially enriched upon Zfp217 knockdown (Figure S5C), providing strong evidence that ZFP217 modulates the m6A RNA levels by counteracting METTL3 activity.
Figure 6. Analysis of ZFP217-dependent m6A sites.
(A) Cumulative distribution function of log2 peak intensity of m6A-modified sites in control and _Zfp217_-depleted ESCs.
(B) Functional categories of ZFP217-dependent m6A sites. The p-value for the enrichment of biological process GO-term is shown.
(C) GSEA plot of ZFP217-dependent m6A sites during RA-induced differentiation (upper panel) and EB formation (lower panel). High and low expression of genes is represented in red and blue, respectively. NES, normalized enrichment score; FDR, false discovery rate.
(D) Heat map representing Log2 fold change of Zfp217 shRNA ESCs compared to control (first column) and Mettl3 KO ESCs compared to WT (second column). Red and blue indicate increase and decrease of m6A peak intensity, respectively.
(E) Distributions of distance between the m6A peak and the nearest consensus site of control and _Zfp217_-depleted ESCs.
(F) Sequence logo representing the consensus motif following clustering of all enriched motifs in ZFP217-dependent peaks.
(G) Venn diagram showing the overlapping transcripts of ZFP217 and METTL3 RIP-Seq samples.
(H) Diagram depicting the coverage at the TSS of genes containing ChIP and RIP peaks or ChIP peaks only with ZFP217 antibodies.
(I) Coverage of ZFP217 signal at the TSS of modified and unmodified genes.
Because ZFP217 was associated with a large number of RNA binding proteins (RBPs), we sought to test the RNA binding capacity of ZFP217 per se. We performed RNA immunoprecipitation coupled with sequencing (RIP-Seq) for ZFP217 and METTL3, and identified 5,933 and 6,971 RNAs associated with ZFP217 and METTL3 (Table S5), respectively. The vast majority of ZFP217-bound transcripts notably overlapped with the METTL3 transcriptome (Figure 6G), and mainly composed of mRNAs associated with transcription and regulation of transcription (Figure s S5D–F). To further explore a connection between the epigenome and the epitranscriptome, we generated enrichment profiles around the genomic region of ZFP217 target genes with ChIP-Seq and RIP-Seq datasets for ZFP217. ZFP217 occupancy at promoters of genes encoding ZFP217-associated transcripts was higher than in non-associated transcripts (Figures 6H and S5G). Moreover, RNAs harboring m6A modification experienced a higher ZFP217 binding, but not POU5F1 binding, at the TSS compared to the transcripts that lacked m6A modification (Figures 6I and S5H). Collectively, these findings indicate that ZFP217 tightly coupled epigenetic regulation and m6A RNA modification. The observation that RNA pol II occupancy was enriched in modified transcripts (Figure S5I) suggests that m6A deposition might occur in parallel with transcription although further validation is required.
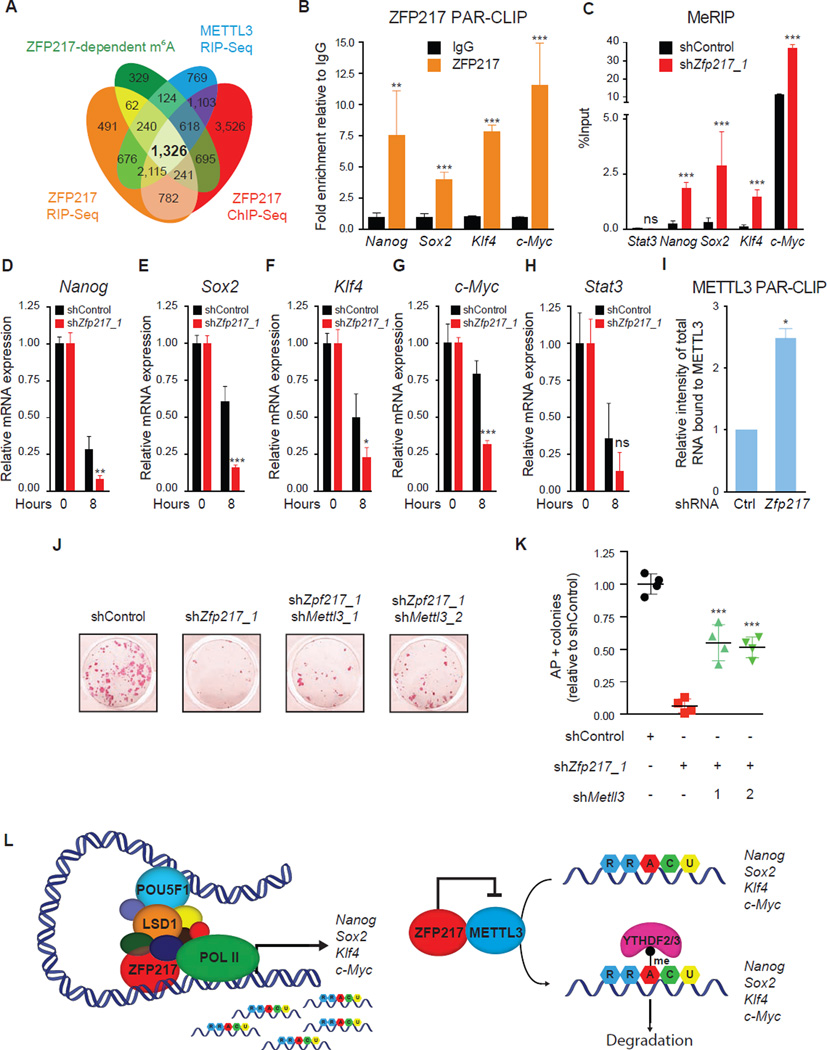
ZFP217 regulates the epitranscriptome of key pluripotency factors affecting somatic cell reprogramming
To further integrate the global mechanism by which ZFP217 regulates the ESC state, we merged ZFP217 ChIP-Seq dataset with m6A ZFP217-dependent transcripts, and ZFP217 and METTL3 transcriptomes (Figure 7A). We identified the core pluripotency factors Nanog, Sox2 (Figure s S6A and S6B), Klf4 and c-Myc, and other factors with a reported function in ESC maintenance (Table S6). Interestingly, Wtap, one of the core components of the m6A methyltransferase complex, and the ‘readers’ Ythdf2 and Ythdf3 were also detected in this group, indicating that ZFP217 might play an active role not just in modulating m6A deposition, but also in modulating the clearance of this RNA modification. To assess the robustness and reproducibility of our analysis, we performed Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP) with ZFP217 antibodies following RT-qPCR. We detected binding of ZFP217 at pluripotency transcripts (Figure 7B), but not at U1, which was used as a negative control. Moreover, MeRIP revealed an increase of m6A modification at these transcripts upon Zfp217 knockdown (Figures 7C and S6C–D), which was associated with a decrease of the lifetime of Nanog, Sox2, c-Myc and Klf4 mRNAs (Figures 7D–G), whereas the stability of the non-target transcripts Stat3 and U1 was unaffected (Figures 7H and S6E). Given that the catalytic domain of METTL3 is also involved in RNA recognition (Bujnicki et al., 2002), we sough to analyze if ZFP217 was affecting METTL3 binding to RNA, thus its m6A methyltransferase activity, by performing PAR-CLIP with METTL3 antibodies in control and _Zfp217_-depleted ESCs. The amount of RNA associated to endogenous METTL3 was significantly increased after loss of Zfp217 (Figures 7I and S6F), suggesting that ZFP217 sequesters METTL3 into an inactive complex.
Figure 7. ZFP217 regulates the epitranscriptome of key pluripotency factors.
(A) Venn diagram of ZFP217 ChIP-Seq target genes, m6A ZFP217-dependent transcripts, and ZFP217 and METTL3 transcriptomes.
(B) RT-qPCR of Nanog, Sox2, Klf4, and c-Myc after PAR-CLIP with ZFP217 specific antibodies. Fold enrichment relative to IgG control. Error bars show ± SD; n = 3. **p<0.01; ***p<0.001 versus IgG control.
(C) RT-qPCR of m6A modification at key pluripotency RNAs in control and Zfp217 shRNA. Percentage of input is shown. Error bars show ± SD; n = 3. ***p<0.0001; ns= not significant versus control shRNA.
(D–H) RT-qPCR analysis of Nanog (D), Sox2 (E), Klf4 (F), c-Myc (G), and Stat 3 (H) expression after 8 hours of EU incorporation in control and _Zfp217_-depleted ESCs. Error bars show ± SD; n = 2. ***p<0.0001; **p<0.001; *p<0.01; ns= not significant versus control shRNA at 8h.
(I) Quantification of the RNA binding ability of METTL3 in control and _Zfp217_-depleted ESCs (from Figure S6E). RNA binding was normalized to the corresponding pull-downed proteins. Error bars show ± SD; n = 2. *p<0.005 versus control shRNA.
(J–K) AP staining of reprogramming MEFs transduced with OSKM in the presence of the indicated shRNA constructs at day 15 (J). Number of AP+ colonies relative to control iPSCs (K). Error bars show ± SD; n = 4. ***p<0.0001 versus Zfp217 shRNA.
(L) Schematics illustrating ZFP217 function in ESC self-renewal and iPSC reprogramming. In our model, ZFP217 directly regulates transcription of key pluripotency and reprogramming factors, including Nanog, Sox2, Klf4 and c-Myc, and promotes their stabilization by preventing them from METTL3-mediated m6A aberrant methylation.
Because loss of Zfp217 led to an increase of global m6A levels in reprogramming MEFs (Figure S6G), we asked if concomitant depletion of Mettl3 was sufficient to rescue the impairment of iPSC reprogramming after Zfp217 knockdown. Consistent with the fact that METTL3 is dispensable for ICM naïve pluripotency established in vivo (Geula et al., 2015), depletion of Mettl3 in reprogrammable MEFs did not decrease the number of AP-positive iPSC colonies (data not shown). Importantly, depletion of Mettl3 partially rescued iPSC reprogramming in _Zfp217_-depleted cells (Figures 7J, 7K and S6H), suggesting that the increment of m6A levels occurring upon Zfp217 knockdown is also a barrier for efficient somatic cell reprogramming.
Collectively, our results show that ZFP217 is required to prevent aberrant methylation of the core pluripotency and reprogramming factors, inhibiting ESC differentiation and promoting efficient iPSC reprogramming. We propose a model in which ZFP217 regulates stem cell maintenance and self-renewal by different mechanisms that are not mutually exclusive and can influence each other (Figure 7L). That is, ZFP217 directly regulates transcription of key pluripotency and reprogramming genes, including Nanog, Sox2, Klf4 and c-Myc, and promotes their stabilization by preventing METTL3-mediated m6A aberrant methylation.
DISCUSSION
Function of ZFP217 in ESC self-renewal and somatic cell reprogramming
ESCs represent an invaluable resource to investigate human disease. However, in-depth understanding of the epigenetic and epitranscriptomic mechanisms controlling self-renewal, pluripotency and transitions to differentiated cell fates is necessary to hold great promise for regenerative medicine. Here, we conducted detailed multidisciplinary research to characterize the role of ZFP217 in ESCs, and we identified ZFP217 to be essential for maintaining the undifferentiated state by tightly coupling epigenetic regulation and m6A RNA modification.
The high Zfp217 expression levels in ESCs and during iPSC reprogramming is a consequence of the direct regulation of ZFP217 by the pluripotency factors NANOG, POU5F1 and SOX2. In addition, we observed severely impaired pluripotency through the downregulation of Nanog, Pou5f1 and Sox2 expression in ESCs depleted of Zfp217, suggesting that ZFP217 is essential in the control of early embryogenesis and it may operate within an autoactivating feedback loop. Moreover, our results show that depletion of Zfp217 in ESCs results in cell proliferation defects by triggering cell cycle arrest and apoptosis, which further shifts the self-renewal phenotype towards differentiation.
In this study we also illustrate the role of ZFP217 in late stages of somatic cell reprogramming when pluripotency factors are activated. Zfp217 depletion significantly decreases both the number of iPSCs and the efficiency of reprogramming, which is in part due to m6A hypermethylation. Three out of four of the Yamanaka factors undergo an increase of m6A of mRNAs upon Zfp217 knockdown, affecting their half-life and the reprogramming capacity. In fact, concurrent depletion of both Zfp217 and Mettl3 is sufficient to partially rescue the impairment of iPSC reprogramming upon Zfp217 loss. These observations could position the modulation of Zfp217 expression as a valuable tool for approaching stem cell therapies.
Comprehensive roles for ZFP217 within an integrated epigenetic regulatory model
Given the broad number of binding regions identified by ZFP217 ChIP-Seq, ZFP217 promoter occupancy is not correlated with changes in gene expression. What is the molecular basis for this promiscuous recruitment in ESCs? ZFP217 DNA-binding activity for specific and non-specific sequences is similar, being the exchange rate between the free and bound protein faster for the nonspecific complex (Vandevenne et al., 2013). Thus, ZFP217 would open genomic DNA with its DNA-binding surface, recruiting additional proteins at such sites, and therefore, facilitating targeting of sequence-specific DNA binding sites among a region of non-specific sites.
Overall, we have made different observations from our comprehensive and integrative epigenomic analysis. First, ZFP217 occupancy is highly enriched at TSSs, overlapping with RNA Pol II Ser-5P, suggesting that ZFP217 could be involved in promoter-proximal pausing in ESCs, thereby facilitating co-transcriptional alternative splicing. Indeed, ZFP217 was found to interact with several RBPs involved in most aspects of RNA metabolism, including RNA processing, maturation, and splicing. Second, ZFP217 occupies actively transcribed genes and, to a lesser extent, ZFP217 can also be located at poised or bivalent promoters of neuroectodermal genes. Third, enhancer and promoter regions of stem cell identity genes are co-occupied by ZFP217, LSD1 and POU5F1. Previously published studies have shown the interaction between LSD1 and ZNF217, where ZNF217 is important for LSD1 recruitment at specific genomic loci (Thillainadesan et al., 2012). On the other hand, most of the POU5F1-associated proteins are transcriptional repressors (Liang et al., 2008), which were also found in different reported ZNF217 interactome analysis, suggesting that these proteins could be part of the same transcriptional complex. Ultimately, our work indicates that ZFP217 is required for direct transcriptional activation of the core pluripotency factors. Given that ZFP217 is a bifunctional transcription factor, it is likely that negative regulation of gene expression through ZFP217 is equally important to maintain pluripotency. Understanding the relative contributions between activation and repression could provide fundamental insight regarding the dynamic control of stemness mediated by ZFP217.
ZFP217 in regulating _N_6-Methyladenosine mRNA methylation
Because METTL3 is a member of the m6A methyltransferase complex that has not yet been characterized, our intent was to resolve the ZFP217-METTL3 interaction from our LC-MS/MS analysis. Importantly, the role of METTL3 in ESCs has only been recently investigated. One model proposes that m6A methylation on developmental regulators blocks HuR binding and destabilizes such transcripts, thereby maintaining pluripotency (Wang et al., 2014b). Other studies postulate that m6A is not required for ESC maintenance but for transition of ESCs to differentiated lineages (Batista et al., 2014). In agreement with the later observation, Mettl3 knockout epiblasts and naïve ESCs are viable at the pre-implantation stage. However, they are unable to exit the naïve state at the post-implantation stage, leading to embryonic lethality (Geula et al., 2015). ZFP217 has crucial regulatory functions in m6A deposition, adding an additional layer of complexity in our understanding of the molecular events regulating RNA methylation in ESCs. ZFP217 sequesters METTL3, diminishing METTL3 binding to RNAs. Moreover, the fact that ZFP217 does not interact with METTL4 strongly suggests that METTL3-ZFP217 is held in an inactive complex. How do the cells dynamically control m6A RNA modification in ESCs? Conceptually, we propose that in the undifferentiated state high levels of METTL3 methyltransferase activity are tightly regulated by ZFP217, preventing core ESC transcripts from aberrant methylation. During cell differentiation, expression of ZFP217 and its target genes rapidly decreases. METTL3 is free and catalyzes m6A methylation at the few pluripotency transcripts that still remain, a requirement to exit the pluripotency state. ZFP217 occupancy at promoters of genes encoding ZFP217-bound transcripts and m6A-methylated RNAs is higher than at unbound and unmodified transcripts, strongly suggesting that ZFP217 controls different regulatory layers of pluripotency, possibly in a redundant and overlapping manner. Therefore, it is reasonable to think that epigenetic mechanisms may impose the composition of the stem cell epitranscriptome.
In summary, we demonstrate that ZFP217 is critical for the maintenance of ESC self-renewal and somatic cell reprogramming by regulating the transcription of pluripotency genes and preventing such transcripts from aberrant m6A methylation. The direct connection between ZFP217 and m6A methylation in maintaining and for reacquiring a pluripotent state provides fundamental insight into the post-transcriptional gene regulation network in stem cell biology. Given the emerging prominence of ZNF217 in oncogenesis (Collins et al., 1998; Littlepage et al., 2012), we suggest that m6A modification is relevant in human cancers. Dissecting the molecular mechanisms that mediate these methylation changes in RNA could predict cancer risk, achieve early diagnosis, track the prognosis of tumor fate, and ultimately, could provide novel therapeutic approaches.
EXPERIMENTAL PROCEDURES
For full details, see the Supplemental Experimental Procedures.
Cell culture and differentiation assays
CCE murine ESCs were grown under typical feeder-free ESC culture conditions. To induce differentiation with retinoic acid (RA), LIF (eBioscience) was removed and RA (Sigma-Aldrich) was added at a concentration of 5 µM. EBs were obtained by growing ESCs in low-attachment dishes in the presence of complete medium and without LIF.
Somatic cell reprogramming
Early passage MEFs were infected with a single lentiviral stem cell cassette (STEMCCA), constitutively expressing all four Yamanaka factors. The day after infection, MEFs were replated at a density of 50,000 cells/ well on irradiated MEF feeder layers and cultured in mouse iPSC media.
ChIP assays
ChIP was performed as previously described (Whyte et al, 2012). The following antibodies were used: anti-ZNF217 (Santa Cruz, sc 55351×), anti-OCT3/4 (Santa Cruz, sc-8628) and anti-LSD1 (Abcam, ab17721).
Analysis of m6A levels
RNA m6A levels were determined by dot blot, m6A immunostaining and LC-MS/MS as described in Supplemental Experimental Procedures. m6A MeRIP-Seq was conducted as previously described (Dominissini et al., 2012).
PAR-CLIP
CCE cells were incubated with 200 µM of 4SU (Sigma Aldrich) for 14 hrs and were crosslinked with 0.4 J cm−2 at 365 nm. After lysis, immunoprecipitation was carried out with ZFP217 or METTL3 antibodies (5 µg and 3 µg, respectively) over night at 4 °C. Precipitated R NA was labeled with [γ-32-P]-ATP and visualized by autoradiography. For PAR-CLIP-RT-qPCR analysis, proteins were removed with Proteinase K digestion, RNA was extracted using Trizol and the RNeasy Mini Kit (Qiagen), and RNA was reverse transcribed with SuperScript® VILO™ cDNA Synthesis Kit. RT-qPCR analysis of the retrotranscribed RNA was performed with specific primers as indicated.
Statistical Analysis
All values were expressed as mean ± SD. Statistical analysis was performed by the Unpaired Student’s t test. A probability value of p < 0.05 was considered statistically significant.
Supplementary Material
1
2
3
4
5
6
7
HIGHLIGHTS.
- ZFP217 regulates the expression of the core stem cell gene network.
- ZFP217 is required for efficient somatic cell reprogramming.
- ZFP217 interacts with METTL3 and restrains m6A RNA modification.
- Low m6A levels in ESC-related transcripts enable pluripotency and reprogramming.
ACKNOWLEDGEMENTS
We thank Dr. A. Alonso of the Weill-Cornell College of Medicine’s Epigenomic Sequencing Core for expert advice for sequencing and library preparation. We thank Dr. Carvajal-Gonzalez (University of Extremadura), Dr. Marrero (University of Barcelona) and Dr. Gacias (Icahn School of Medicine at Mount Sinai) for critically reading the manuscript, and D. Munoz for graphical design. F.A. is supported by the Catalan Agency for Administration of University and Research (AGAUR) under a Beatriu de Pinos postdoctoral fellowship. This research was supported by a Senior Scholar Award in Aging (AG-SS-2482-10) to M.J.W. from the Ellison Medical Foundation and awards HL103967, DA028776 and CA154903 from the NIH to M.J.W. J.A.W. was supported by award GM089778 from the NIH. J.W. was funded by grants from the NIH (1R01-GM095942) and the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM C028103 and C028121).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information includes six figures, eleven tables, experimental procedures and supplemental references.
AUTHOR CONTRIBUTION
F.A. and M.J.W. conceived and designed the project. F.Z. and A.S. contributed equally to this work. F.Z. and W.Z. performed all bioinformatics analysis except for ChIP-Seq that was also analyzed by B.A. and A.R. F.A., A.S., M.F., S.D.C., A.V., D-F.L., C-H.C., M.R., F.J., R.W., J.A.W., and J.W. performed experiments. S.R.K. provided with important reagents. F.A., A.S., and M.J.W. wrote the manuscript.
REFERENCES
- Banck MS, Li S, Nishio H, Wang C, Beutler AS, Walsh MJ. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics. 2009;4:100–106. doi: 10.4161/epi.4.2.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m(6)A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, et al. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci U S A. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–3386. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, Feuerstein B, Mills GB, Stokoe D, Yaswen P, et al. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum Mol Genet. 2005;14:3219–3225. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krig SR, Jin VX, Bieda MC, O'Geen H, Yaswen P, Green R, Farnham PJ. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Li P, Maines-Bandiera S, Kuo WL, Guan Y, Sun Y, Hills M, Huang G, Collins CC, Leung PC, Gray JW, et al. Multiple roles of the candidate oncogene ZNF217 in ovarian epithelial neoplastic progression. Int J Cancer. 2007;120:1863–1873. doi: 10.1002/ijc.22300. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Littlepage LE, Adler AS, Kouros-Mehr H, Huang G, Chou J, Krig SR, Griffith OL, Korkola JE, Qu K, Lawson DA, et al. The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov. 2012;2:638–651. doi: 10.1158/2159-8290.CD-12-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet GH, Stampfer MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001;61:1250–1254. [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, Bolognesi M, Crossley M. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006;26:8159–8172. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan KG, Verger A, Yaswen P, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim Biophys Acta. 2007;1775:333–340. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thillainadesan G, Chitilian JM, Isovic M, Ablack JN, Mymryk JS, Tini M, Torchia J. TGF-beta-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Molecular cell. 2012;46:636–649. doi: 10.1016/j.molcel.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Thollet A, Vendrell JA, Payen L, Ghayad SE, Ben Larbi S, Grisard E, Collins C, Villedieu M, Cohen PA. ZNF217 confers resistance to the pro-apoptotic signals of paclitaxel and aberrant expression of Aurora-A in breast cancer cells. Mol Cancer. 2010;9:291. doi: 10.1186/1476-4598-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck MT. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int J Biochem. 1992;24:379–386. doi: 10.1016/0020-711x(92)90028-y. [DOI] [PubMed] [Google Scholar]
- Vandevenne M, Jacques DA, Artuz C, Nguyen CD, Kwan AH, Segal DJ, Matthews JM, Crossley M, Guss JM, Mackay JP. New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217. J Biol Chem. 2013;288:10616–10627. doi: 10.1074/jbc.M112.441451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell JA, Thollet A, Nguyen NT, Ghayad SE, Vinot S, Bieche I, Grisard E, Josserand V, Coll JL, Roux P, et al. ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 2012;72:3593–3606. doi: 10.1158/0008-5472.CAN-11-3095. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welham MJ, Kingham E, Sanchez-Ripoll Y, Kumpfmueller B, Storm M, Bone H. Controlling embryonic stem cell proliferation and pluripotency: the role of PI3K- and GSK-3-dependent signalling. Biochem Soc Trans. 2011;39:674–678. doi: 10.1042/BST0390674. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HB, Johnson R, Kunarso G, Stanton LW. Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 2011;21:1284–1293. doi: 10.1101/gr.114488.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7