Mining the human gut microbiota for immunomodulatory organisms (original) (raw)
. Author manuscript; available in PMC: 2020 Dec 31.
SUMMARY
Within the human gut reside diverse microbes coexisting with the host in a mutually advantageous relationship. Evidence has revealed the pivotal role of the gut microbiota in shaping the immune system. To date, only a few of these microbes have been shown to modulate specific immune parameters. Herein, we broadly identify the immunomodulatory effects of phylogenetically diverse human gut microbes. We monocolonized mice with each of 53 individual bacterial species and systematically analyzed host immunologic adaptation to colonization. Most microbes exerted several specialized, complementary, and redundant transcriptional and immunomodulatory effects. Surprisingly, these were independent of microbial phylogeny. Microbial diversity in the gut ensures robustness of the microbiota’s ability to generate a consistent immunomodulatory impact, serving as a highly important epigenetic system. This study provides a foundation for investigation of gut microbiota–host mutualism, highlighting key players that could identify important therapeutics.
INTRODUCTION
The mammalian gastrointestinal tract is inhabited by hundreds of species of symbiotic microbes, many of which have a beneficial impact on the host (Kamada et al., 2013; Kau et al., 2011). The local immune system faces the daunting task of enforcing peaceful co-existence with these microbes while also imposing a staunch barrier to pathogen invasion (Hooper and Macpherson, 2010). Maintaining this equilibrium involves both the innate and adaptive arms of the immune system as well as non-immunologic protective strategies—e.g., those involving the mucus barrier and antimicrobial peptides (AMPs) (Hooper et al., 2012). These host-protective mechanisms are counterbalanced by regulatory processes that limit the antibacterial response and prevent collateral damage from inflammation.
The gut microbiota plays an important role in educating and modulating the host immune system (Mazmanian et al., 2005, Schirmer et al., 2016). Germ-free (GF) mice show defects in multiple specific immunocyte populations, such as Th2 skewing of their CD4+ T cell compartments, compromised innate lymphoid cell (ILC) function; a deficiency in IgA-producing plasma cells; and, more generally, greater susceptibility to infection (Surana and Kasper, 2014; Hepworth et al., 2013).
The immunologic impacts of few microbial species have been elucidated: Segmented Filamentous Bacteria (SFB) elicit a robust Th17 response (Ivanov et al., 2009; Gaboriau-Routhiau et al., 2009); a glycosphingolipid from Bacteroides fragilis inhibits invariant natural killer T cell differentiation (An et al., 2014); and specific subsets of CD4+Foxp3+ regulatory T cells (Tregs) are induced by a range of individual or groups of microbes (Sefik et al., 2015; Faith et al., 2014; Lathrop et al., 2011; Atarashi et al., 2013). These changes in immunocyte profiles have readily discernible effects on both gut and extra-gut immune responses, whether protective or pathogenic. Thus, there has been great interest of late in harnessing immune system–microbiota cross-talk in the intestine to therapeutic ends. A common approach has been to perform microbiome-wide association studies to search for correlations between particular microbes and particular disease conditions (Kostic et al., 2015; Gevers et al., 2014; Smith, 2015). While this is, of course, a useful strategy, we decided to take a different approach. Our driving concept was that the co-evolution of the intestinal microbiota and the local immune system for millennia has resulted in a variety of presumably innocuous strategies by which various microbes manipulate immune system activities. Our goal was to begin to uncover these microbial tactics, using a compendious and performant screen.
The approach we chose was gnotobiotic colonization of GF mice with single microbial strains derived from the human gut followed by extensive immunophenotyping and transcriptomic analysis. While this unabashedly reductionist experimental strategy sets aside the combinatorial effects of a complex microbiota, monocolonization renders the complexities of immune system–microbiota interactions more tractable. The numbers of colonizing bacterial species are higher and more stable over time in a monocolonized host than in a host with a diverse microbiota, and the antigenic or metabolic stimulus to the local immune system is consequently stronger. We were seeking not to recapitulate the biology of the human gut, but rather to establish a robust, “sensitized” readout system that permits screening for human-derived immunomodulatory microbes and molecules. Our screen focused on human intestinal symbionts that encompassed, as widely as was practical, the genetic diversity of the human gut microbiota.
Ultimately, we monocolonized mice with 53 individual bacterial species representing all five of the major phyla, and we evaluated their effects on the composition and activation of most innate and adaptive immune-system cell types as well as on intestinal tissue transcriptomes. We present both a synthetic overview of the extensive dataset generated and three vignettes describing our findings on particular immunomodulatory cell types or molecules.
RESULTS
We set up a systematic screen for immunomodulatory human gut symbionts. GF C57BL/6 mice were bred in an isolator under rigorous microbial monitoring. At 4 weeks of age, eight mice were sterilely transferred to a gnotobiotic isolator, where they were colonized by gavage with one of the study’s 62 bacterial strains (Table S1A). Fifty-three species spanning the known human gut diversity were originally selected for complete analysis; 9 additional strains were chosen for focused analysis to determine whether interesting findings were shared across a species. Two weeks after colonization, host response was assessed by immunologic and genomic profiling of colon and small intestine (SI) (Fig. 1A). Six week old GF mice were regularly analyzed throughout. Standard operating procedures were strictly followed. All experiments included were documented to ensure monocolonization only with the desired microbe by culture and 16S rDNA sequencing. Moreover, feces from fourteen randomly chosen experiments were analyzed by deep sequencing and shown to be pure. We have reported all experiments that were documented to be free of contamination. Phenotypes of interest were validated by independent repetition.
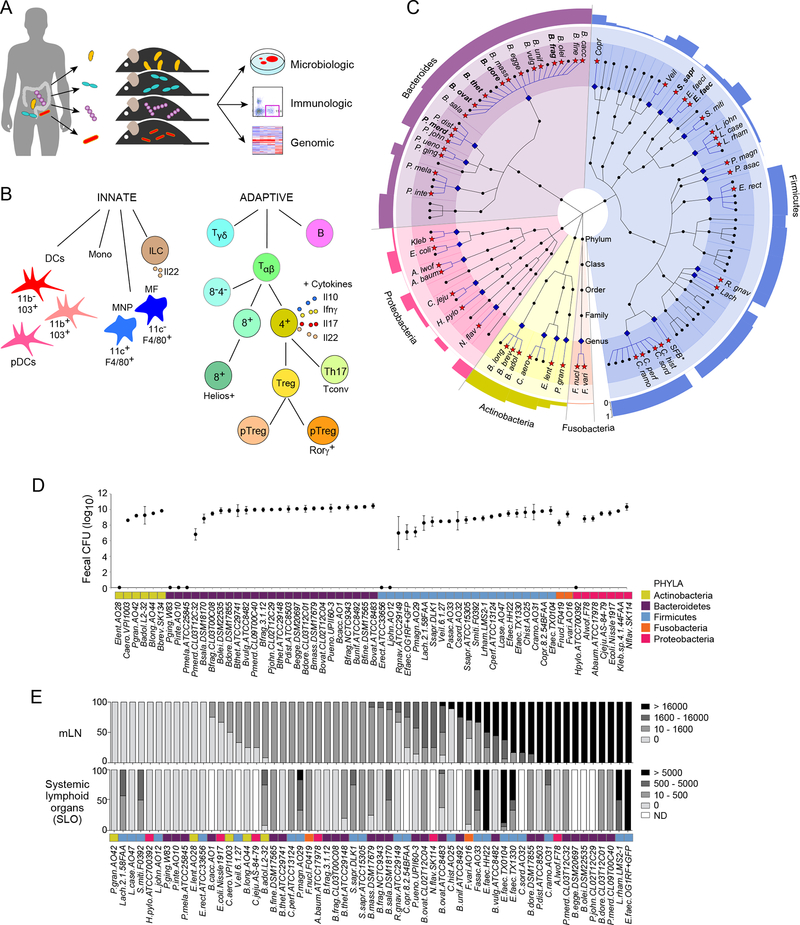
Figure 1: Experiment design and bacterial colonization.
(A) Four week-old GF mice were monocolonized with human gut bacteria and analyzed after 2 weeks for colonization, impact on immune system and genomic activity.
(B) Innate and adaptive immune responses were analyzed by flow cytometry of cells extracted from SI, PPs, colons, mLNs, and SLOs.
(C) Cladogram of the human gut microbiota. Microbes were identified in the Human Microbiome Project (HMP) database except for SFB. Blue diamonds denote the genera included; red stars mark the species. Species where more than one strain was analyzed are in bold type. The outer ring represents a bar graph of the prevalence of each genus.
(D) Average CFU per gram of feces, and their standard deviations.
(E) Bar graphs of CFUs in mLNs (per organ, top) and SLO (bottom).
Both local and systemic effects on the immune system were examined by analyzing the proportions of 18 cell types from its innate and adaptive arms (Fig. 1B, Fig. S1, Table S1B). Five intestinal and lymphoid tissues were examined: SI and colonic lamina propria, Peyer’s patches, mesenteric lymph node (mLNs) and systemic lymphoid organs (SLO; pooled spleen and subcutaneous lymph nodes). CD4+ T-cell production of the cytokines Il10, Il17a, Il22, and IFNγ, and ILC production of Il22 were also measured.
Microbial selection and colonization
Fifty-three bacterial species were selected from the Human Microbiome Project to represent the spectrum of phyla and genera in the human gut microbiota, and covering the 5 dominant phyla: Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria, and Fusobacteria (Fig. 1C, Table S1A).
Effective gastrointestinal colonization was assessed by culture of fecal material harvested from the colon and, in some cases, from the stomach and oral cavity. Most of the strains introduced orally into GF mice successfully colonized the intestines of the recipients (108 to 1010 CFU/g; Fig. 1D; Table S1C). Of the seven species not recovered in fecal specimens, five were recovered at other sites. Porphyromonas gingivalis, Prevotella intermedia, and Prevotella melaninogenica were found only in the oral cavity, while Helicobacter pylori and Lactobacillus johnsonii resided exclusively in the stomach. Interestingly, these are the anatomic sites in which these species are normally found in mice and humans with a complex microbiota. This existence of niche preferences even in the absence of microbial competition suggests that they derive from organ-specific physical and/or chemical properties that are intrinsically unfavorable for a certain microbe, such as acidity or the availability of particular nutrient types, rather than from competitive fitness. Only two bacteria failed to colonize any site (Eubacterium lentum and Eubacterium rectale).
Commensal bacteria can breach intestinal barriers and can be found in small numbers in gut-draining lymph nodes or systemically (Vaishnava et al., 2008). This microbial delocalization is facilitated by deficiencies in innate defenses (Vaishnava et al., 2008; Knoop et al., 2015; Wang et al., 2016) and by myeloid cells that actively transport the bacteria, plausibly to enable antigen presentation (Macpherson and Uhr, 2004; Diehl et al., 2013). Because the ability of various symbionts to partake in extra-intestinal delocalization is unknown, we took advantage of this screen to investigate the ability of the bacteria we studied to delocalize to mLNs and caudal lymph nodes (cLNs), which drain the SI and the colon, respectively, and to the SLO. Strict precautions were taken during dissection to avoid contamination from the gut. A majority (88%) of the species that colonized the gut were detected alive in mLNs (Fig. 1E, top), with no particular preference according to phylum, genus, or aerobe/anaerobe status. A substantial proportion (47%) of gut-colonizing microbes were found alive in the SLO (Fig. 1E, bottom).
Immunologic changes in response to monocolonization with human gut symbionts
The broad screen described above generated 24,255 individual immunophenotypes induced in local or systemic lymphoid organs by the bacteria that successfully monocolonized GF mice and for which complete data were obtained. Fig. 2A and Table S2A illustrate the changes in frequencies of immunocyte populations in the colon, highlighting significant changes at a False Discovery Rate (FDR) of ≤0.01. The corresponding Fold Changes (FCs) relative to GF status are summarized in the heat map in Fig. 2B and Table S2B; results in other tissues in Fig. S2A and Table S2A–C; individual mouse data in Table S2D. A patchwork of effects was observed. Some innate cell types varied in response to several microbes, with expansion (e.g., CD103+ dendritic cells [DCs]), contraction (e.g., both CD11b+F4/80+ subsets of macrophages and mononuclear phagocytes), or both (e.g., plasmacytoid dendritic cells [pDCs]). Type 3 ILCs (ILC3s) were affected by only a few microbes, a result consistent with earlier studies reporting microbiota-mediated alterations in Il22 production but not in overall ILC3 frequency (Lee et al., 2011; Reynders et al., 2011). Most cells of the adaptive immune system seemed largely unresponsive, at least in terms of abundance, with comparatively infrequent and modest changes in the proportions of B, γδT, and αβT (T4 or T8) cells. The notable exceptions were Tregs and their subsets, which, in line with previous reports (Lathrop et al., 2011; Faith et al., 2014; Sefik et al., 2015), were strongly induced by a number of individual microbes. These effects were distributed among the different microbes tested, with a range in the number of cell types affected by a given microbe (as judged by the proportion of cell types modified by a z-score of ≥2 relative to GF; Fig. 2C). Some microbes seemed stealth-like, affecting few or none of the immunocyte populations examined (e.g., Peptostreptococcus magnus and Bacteroides salanitronis), but others were substantially more active (Bacteroides uniformis). Microbes of the same phylum or genus provoked no obviously shared patterns of these signatures in terms of either the number of cell types affected (Fig. 2C) or the extent of change relative to GF (Fig. 2B, Table S2B).
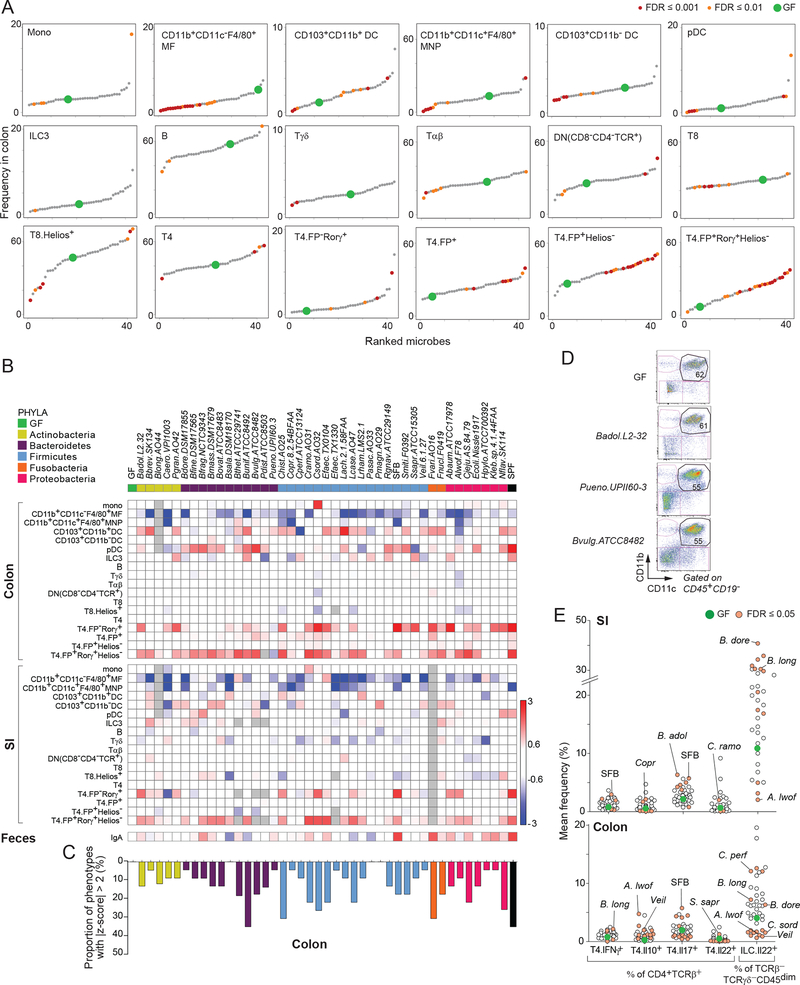
Figure 2: Immunomodulation by gut microbes.
(A) Average frequencies of each immunocyte population for every microbe. For cell type frequency determination (y-axis) and microbe identification (x-axis) see Tables S1A,B, S2A,B, and Fig. S1.
(B) Heatmap of average fold changes (relative to GF) for cells in the colon and SI following monocolonization, and fecal IgA. Gray- no data.
(C) Proportion of colonic immune cell types (compared to GF) with a z-score ≥ 2.
(D) Example of colonization influencing the gating configuration but not frequency of cell populations. Flow cytometry plots shown are for CD11b+CD11c+ MNPs and DCs.
(E) Cytokine responses in SI and colon.
In addition to quantitative changes, we observed with a few microbes some reproducible alterations in the configuration of cell populations within flow cytometry counting gates, as illustrated by the difference in CD11c intensity in CD11b+CD11c+ mononuclear phagocytes and DCs (Fig. 2D; Figs. S2C–E, Table S2C). These changes occurred independently of the quantitative perturbations measured above. We assessed the induction of inflammatory or suppressive cytokines by CD4+ T cells and ILCs; because the staining panels were designed before defined markers for ILC subsets had been established, we assessed only bulk ILC populations (Fig. 2E). Only a handful of symbionts elicited deviations from GF levels in T cells, including (as expected) SFB and Th17 cells (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009), but other unprecedented associations were found, such as Coprobacillus with Il10+ SI T cells and Bifidobacterium longum with colonic Th1 (T4.IFNγ+) cells (Fig. 2E). Bacterial influences on Il22 production by ILCs were far more pronounced, with significant induction by microbes such as Bacteroides dorei and B. longum in both gut tissues. Conversely, Acinetobacter lwoffii, Clostridium sordellii, and Veillonella appeared to repress Il22 production, especially in the colon—a result indicating that the microbes can have differential effects on ILC activation. These observations provide a nuanced perspective on bacterial modulation of ILCs and may explain discrepancies in studies comparing Il22 production in GF and specific pathogen–free (SPF) mice (Satoh-Takayama et al., 2008; Lee et al., 2011; Sawa et al., 2011).
Fecal IgA was quantitated at the end of the 2-week monocolonization. All IgA levels ranged between GF and SPF. Fold change relative to GF is shown in Fig 2C. IgA induction varied by organism and did not follow microbial phylogeny. Total IgA was measured in fecal samples by ELISA and organism-specific IgA was evaluated by flow (Fig. S2F). There was a significant correlation between total and organism specific IgA (_r_=0.51, _p_=0.025). This suggests microbes induce IgA production by acting as standard “immunogens” rather than as bystanders that boost IgA production without being direct targets themselves.
Further insight was obtained by correlating the responses induced by the set of microbes in the colon versus the SI (Fig. 3A). Many of the stronger correlations corresponded to the same cell type in the colon and SI (e.g., F4/80+ mononuclear phagocytes, Il10-producing CD4+ T cells, or RORγ+ Tregs), an observation denoting similar responses despite differences in tissue organization and microbial load in these two gut segments. Other correlated phenotypes, although expected (e.g., ILC3 frequency and the proportion of Il22 producing cells among bulk ILCs; CD4+RORγ+ T cell frequency and Il17a production), did reinforce the significance of the trends observed. Finally, some correlated traits were less anticipated (e.g., Tγδ and Helios+CD8+ T cells; CD4+ T and B lymphocytes) and may reflect common sensing pathways or integration of microbial influences by the immunologic network.
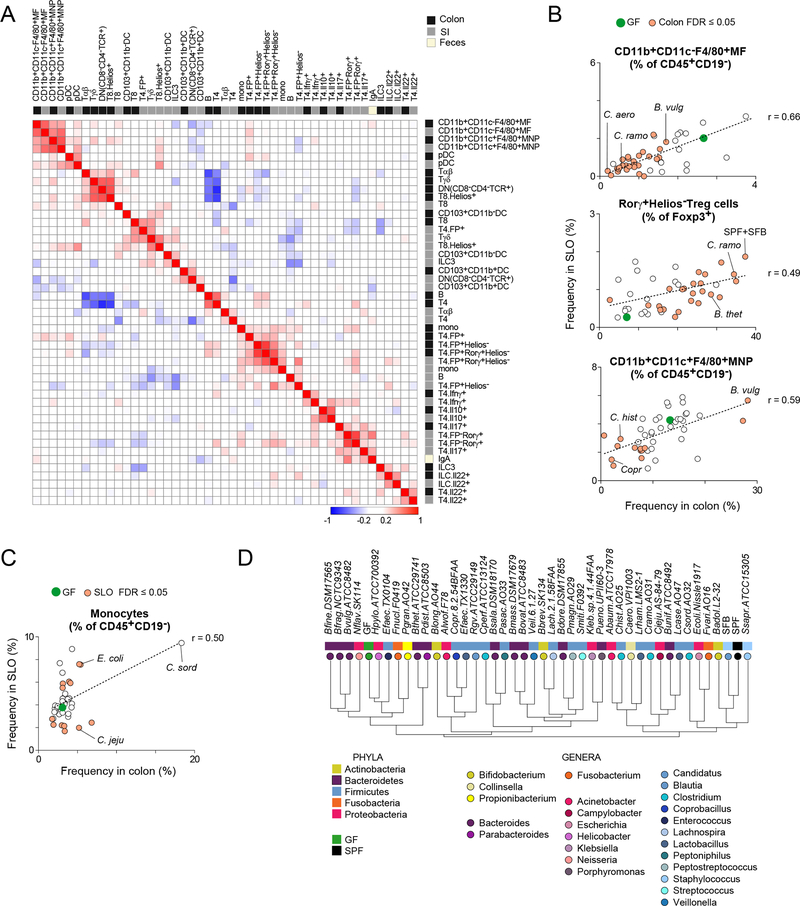
Figure 3: Local and systemic immunologic correlations.
(A) Clustered heatmap of Pearson correlation coefficients (r) for immunophenotypes after moncolonization.
(B) Average cell frequency correlations: SLO versus colon, for MFs(upper), Tregs (middle) and MNPs (lower).
(C) Average cell frequency correlations: SLO versus colon, for monocytes.
(D) Hierarchical clustering dendrogram of bacteria based on the Pearson correlation of their overall immunologic impact on the SI and colon. Values were normalized to the mean across all microbes.
See also Figure S3.
Bacteria of the same phylum or genus provoked no obviously shared patterns of signatures in terms of either the number of cell types affected (Fig. 2C) or the extent of change relative to GF (Fig. 2B, Table S2B). We correlated the normalized immunophenotypic responses between microbes in the SI and the colon (Figs. 3D and S3B). The dendrogram generated by hierarchical clustering of these correlations bore testament to the true diversity of microbial functions represented by the organisms chosen for this screen. Bacterial species from the same phylum or genus largely failed to cluster together, a result pointing to a high degree of diversification in immunomodulatory properties within a phylum or genus. For seven species (nine strains total), we looked at the impact of additional strain(s) on lymphocyte populations such as Tregs. For the Bacteroides strains within the same species, we found quantifiable differences (Table S2D). The mean Euclidean distance between species was 0.39. Interestingly, the mean distance between strains within the same species was very similar- 0.32. These results highlight the importance of strain-level information in relating microbial function to immunologic phenotypes.
Effects of bacterial colonization in systemic lymphoid organs
Immunocytes can migrate from the colon into the lymphatics and circulate between lymphoid organs (Morton et al., 2014). We analyzed immunocyte populations in the mLNs and the SLO to determine whether immunologic alterations in the gut were reflected systemically. Most microbes had a limited effect on innate immunocytes in mLNs and the SLO (Fig. S2B), although monocytes did vary markedly in the SLO. As in the intestine, adaptive immunocytes in lymphoid organs were mostly unaffected by microbial exposure. To detect more sensitively the echoes in lymphoid organs of microbe-instructed immunologic changes in the gut, we correlated the immunologic phenotypes in the gut and secondary lymphoid organs (Figs. 3B, and S3A). There was a significant correlation across all tissues for five cell types. For three of these types (the F4/80+ macrophage and mononuclear phagocyte populations and FoxP3+ Tregs), changes in the SLO were subtle but were correlated with frequencies in the gut across the set of microbes (Fig. 3B). This finding suggested a direct relationship between the two pools. The fifth cell type—the monocyte—was the exception, with equally strong induction by C. sordellii in the SLO and the intestines (Fig. 3C).
Colonic and SI transcriptomes of monocolonized mice
We next investigated the transcriptomic changes induced by the various microbes in SI and colonic tissue. Gene-expression profiles were generated in duplicate from whole-tissue RNA in order to capture responses in all major cell types, with controls from GF tissues included in every batch. We found more marked inter-individual variability in intestinal tissues than in other tissues we have recently profiled such as the fat and muscle (data not shown). Groups of variable genes appeared in the plot of gene-wise coefficients of variation (CV) (Fig. 4A): one group had the same variability in replicates of GF and monocolonized mice, but a larger group was more variable in GF colons than in microbially colonized colons, as if the presence of bacteria stabilized fluctuations in the transcriptome. Except for some B cell–specific transcripts, most of these highly variable genes could not be ascribed to fluctuations in the frequency of particular cell types.
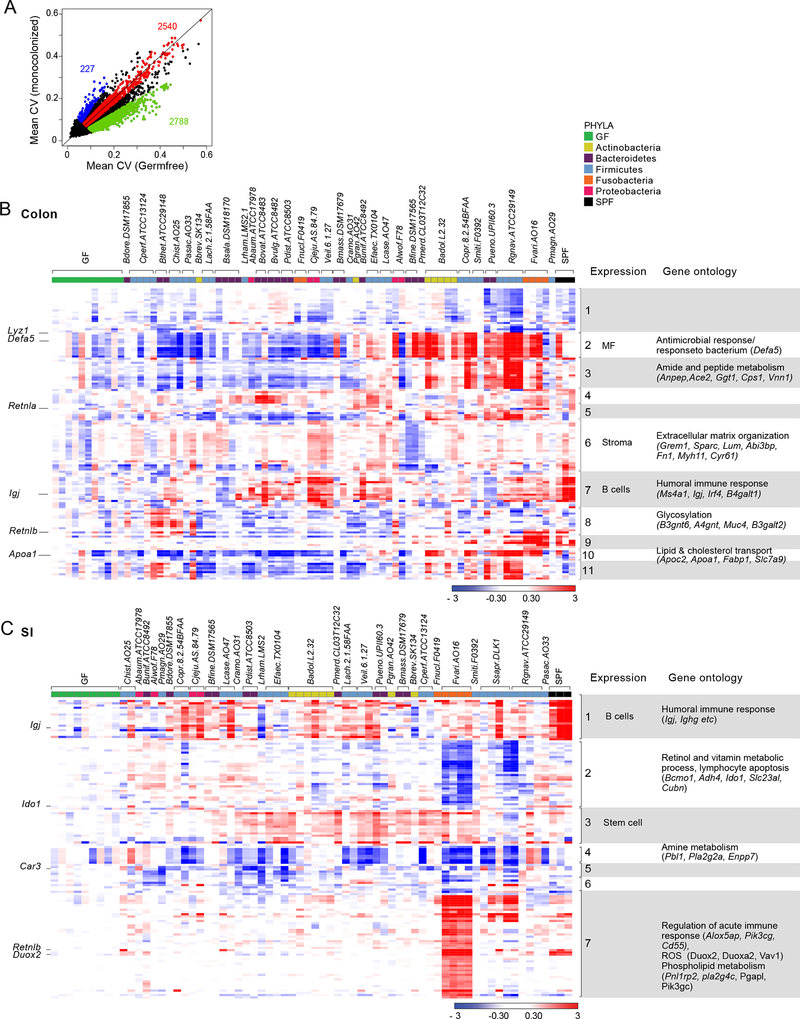
Figure 4: Transcriptional responses to colonization.
(A) Mean coefficient of variation (CV) in transcripts from the colons of monocolonized mice and GF mice.
(B-C). Heatmap of fold changes of transcripts differentially expressed in (B) the colon and (C) SI of monocolonized and SPF mice compared to GF mice. Bacteria (columns) are clustered by hierarchical clustering; Genes (rows) are clustered by K-means clustering. Association of these transcripts with particular immune and non-immune cell types was verified in gene expression databases such as ImmGen and GNF. Enriched pathways were identified using GO.
This degree of background variation made the determination of microbe-specific effects somewhat more complicated, but clear effects were apparent in volcano plot representations (Figs. S4A and S4B). We opted for a general approach, flagging transcripts with an FC relative to GF >2.5 (or <0.4) and uncorrected p(-log10)>2.5 for at least one bacteria. This selection yielded an unexpectedly small number of transcripts, indicating that symbiotic bacteria have only limited effects on the gut transcriptome in the monocolonization setting: 128 genes were up- or down-regulated in the colon and 116 in the SI, of which 20 were responsive in both colon and SI (Table S3). These transcripts are displayed for each microbe in Figs. 4B and 4C. None of them was uniformly induced by all bacteria, but >60% of these responsive transcripts were induced by some microbes and repressed by others (e.g., Defa5, Retnlb, Apoa1, and Lyz1 in the colon_; Retnlb, Duox2,_ and Reg3a in the SI). This observation indicated that different microbes can sometimes have diametrically opposed consequences. Some bacteria may take advantage of the host’s adaptive abilities as a means of out-competing other microbes, either by creating a more favorable environment for themselves (Rakoff-Nahoum et al., 2016) or by down-regulating host metabolic pathways such as those for lipid or amine metabolism to create a hostile environment for other bacteria that require these molecules.
Some bacteria had stronger and more reproducible signatures (e.g. Fusobacterium varium in the SI, Campylobacter jejuni in the colon), while others had weaker and more variable imprints (Bacteroides salanitronis, Clostridium perfringens). None of the transcripts was uniquely induced by a single microbe, but most were induced (or repressed) by several bacteria, with no particular connection to phylum. In these respects, the diversity of transcriptional changes mirrored the alterations in immunophenotypes described above. These transcriptomic changes could be grouped in co-regulated gene clusters (Figs. 4B and 4C). Cross-referencing to gene-expression databases (ImmGen, GNF atlases) showed that some, but not all, of these clusters were predominantly expressed in particular cell types and probably corresponded to responses in those cells (e.g., stromal, macrophage, B cell, or perhaps even stem cell transcripts; Figs. 4B and 4C). In both tissues, the responsive genes encoded a variety of functional molecules—AMPs, stress response elements (Retn, Retnla, Retnlb), hemoglobins (likely reflecting changes in vascularization), immunoglobulin-related transcripts, and enzymes and molecules involved in lipid metabolism (fat digestion and absorption, lipid processing, lipase and phospholipase activity)—with corresponding overrepresentation of Gene Ontology pathways (antimicrobial response, extracellular matrix organization, amide and amine metabolism, retinol and vitamin metabolism, and acute inflammatory response). There was also an enrichment in transcripts reported to be affected in infant mice secondary to maternal colonization (Gomez de et al., 2016). On the other hand, we did not observe significant induction of inflammation-associated cytokines like Il1α, Il1β, Il6, Il22, TNF, Il12, or IFNs. (Levels of Il1a, Il22, and Il6 were below detection.) However, Il18 levels were slightly elevated in response to several different bacteria (Figs. S4C and S4D).
Immunomodulatory cell types and transcriptional responses
Some interesting observations that we think merit emphasis are presented.
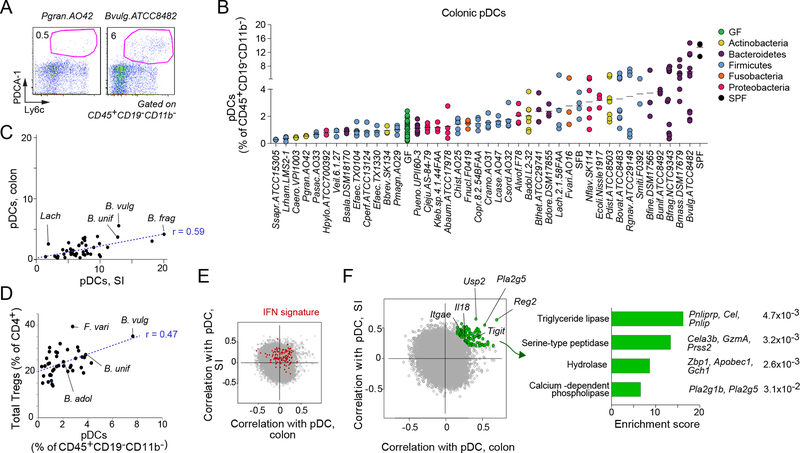
Colonic pDCs are biased by gut bacteria.
Plasmacytoid dendritic cells are distinctive players in the innate arm of the immune system, playing a central role in antiviral defenses through their ability to produce copious amounts of type I IFNs. Correspondingly, they have been implicated in several IFN-linked diseases (Swiecki and Colonna, 2015). The influence of the gut microbiota on the pDC pool is largely unknown. One report described a reduction in pDCs in mice with a restricted microbiota distinct from that typical of SPF mice (Fujiwara et al., 2008), while another study revealed induction of pDCs in mLNs by B. fragilis during ongoing colitis (Dasgupta et al., 2014). Among the myeloid populations, pDCs had the greatest range of fluctuation in our screen (Fig. 2A), as exemplified by the cytofluorometry profiles in Fig. 5A. These fluctuations were bidirectional (Fig. 5B): 38% of the bacteria tested increased colonic pDC proportions (by ≥2-fold) in monocolonized mice over those in GF mice, while 8% reduced colonic pDC proportions by >2-fold—most extremely in mice colonized with Staphylococcus saprophyticus and Lactobacillus rhamnosus, which harbored almost no pDCs. However, these frequencies were quite variable even in mice colonized by the same organism. For instance, Bacteroides vulgatus (ATCC 8482) was the most potent species at inducing colonic pDCs on average (mean, 6.4% pDCs), but with a range from 1.7% to 14.7%. The recalibration of pDCs in the colon resulting from monocolonization was more variable than the recalibration of CD103+ DCs in the same mice (Fig. S5). Interestingly, the ability of a microbe to induce pDCs in the SI and the colon was significantly correlated (_r_=0.52, _t_-test _p_=0.0006l; Fig. 5C); this correlation suggests that the same mediators or pathways might be at play in the two organs. pDCs have significant tolerogenic potential and can stimulate Tregs (Reizis et al., 2011), an ability that has been associated with type I IFN production (Metidji et al., 2015; Kole et al., 2013; Nakahashi-Oda et al., 2016). Also of interest was the significant correlation between the strains’ ability to boost colonic pDCs and total FoxP3+ Treg frequencies (_r_=0.46, _t_-test _p_=0.003; Fig. 5D).
Figure 5: Colonic plasmacytoid dendritic cells are most prolific myeloid responders to the gut microbiota.
(A) Representative flow cytometry dot plots of a pDC ‘low inducer’, Propionibacterium granulosum (Pgran.A042) and a ‘high inducer’ B. vulgatus (Bvulg.ATCC8482). Cells were gated as CD45+CD19-CD11b-.
(B) Frequencies of pDCs in the colon induced by monocolonization
(C) Pearson correlation between pDC’s in SI vs. colon (_p_=0.0006).
(D) Pearson correlation between colonic pDCs and Tregs (_p_=0.003).
(E-F) Correlation coefficients were calculated between the expression value of each gene from the whole tissue transcriptome (SI, and colon) and the proportions of pDCs for each monocolonizing microbe (SI and colon). (E) Genes related to the interferon signature are marked in red. (F) Genes having similar expression patterns and correlating best in both SI and colon are highlighted in green. (Right) Bar graph of the enriched biological pathways of these highly correlating genes as analyzed by Enrichr. Most significant pathways determined by GO Molecular Function (p<0.05). Depicted gene names and the actual Enrichr adjusted p.values are shown.
Next, we identified the sets of genes whose expression was most correlated with pDC frequencies in the SI or the colon, information that we thought might yield insights into the molecular pathways through which microbes modulate pDCs and/or the physiological consequences of their pDC levels. No clear cluster of outliers stood out in these correlations. However, a set of IFN-inducible signature transcripts showed an enhanced correlation with pDC frequencies in both the SI and the colon (Fig.5E, red dots), which was likely a reflection of their characteristically abundant IFN production. This set of genes (Fig. 5F, left panel, green dots; Table S4) included a few interesting transcripts worth highlighting. One transcript, Il18, was noteworthy given that pDCs express high levels of Il18R2 and that Il18 antagonizes their production of type I IFN. One might speculate that Il18 induced by some microbes can promote pDC accumulation rather than effector function (Chao et al., 2014). Another transcript was Tigit, an activation marker on T cells whose particular expression on Tregs may relate to the correlation between pDC and Treg proportions. Overall, the transcripts most correlated with pDC frequency were enriched in lipid or protein digestion and metabolic pathways (Fig. 5F, right panel), an observation suggesting a connection between pDCs and the metabolic and nutrient uptake functions of the gut.
Antimicrobial peptide expression upon microbial colonization.
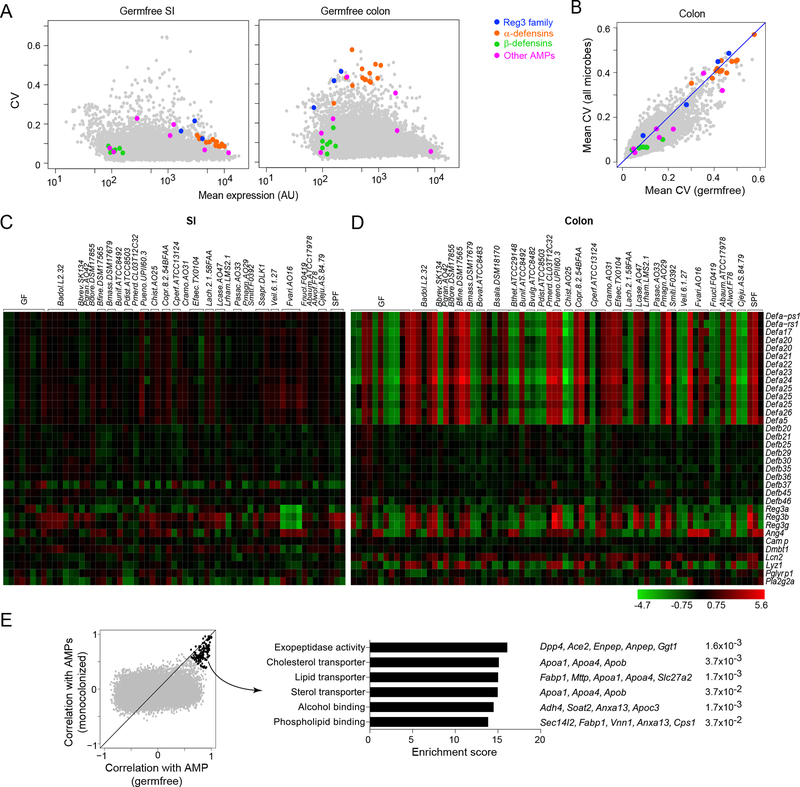
Expression of many gut AMPs is constitutive, although bacterial colonization can induce a subset of these peptides in SI Paneth cells (Bevins and Salzman, 2011; Gallo and Hooper, 2012). We asked whether AMPs respond similarly to different bacterial species and whether they are coordinately regulated in the SI and the colon. In GF mice, α-defensins, Reg3 family members, and other Paneth cell–derived products (such as Ang4) were expressed at reproducibly high levels in the SI but at 20-fold lower levels in the colon, (Fig. 6A), where they were among the most variably expressed transcripts genome-wide (as indicated by their reproducibly high CV, Fig. 6A–B) In contrast, β-defensins, which are produced by many types of epithelial cells (Bevins and Salzman, 2011; Gallo and Hooper, 2012), were expressed at comparable levels in the SI and the colon.
Figure 6: Antimicrobial peptides exhibit divergent patterns of expression in the SI and colon.
(A) Coefficient of variation (CV) vs. mean expression in GF mice for all genes in the SI (left panel) and colon (right panel). Only genes expressed above background level are shown. Antimicrobial peptides (AMPs) are highlighted and color-coded according to the categories listed.
(B) The CV of all expressed genes in the colons of GF vs monocolonized mice, as shown in Fig. 4A, here AMP genes highlighted.
(C-D) Heatmaps illustrating the differential expression of AMPs in the SI (C) and colon (D) in various monocolonized mice compared to GF. Heatmap colors represent the log2 fold change values relative to GF. Only AMPs expressed above background levels are shown.
(E) Gene programs correlated with AMP expression in the colon. For every gene expressed in the colon, its correlation with colonic AMP genes (Reg3 family and α-defensins) is plotted for GF vs. monocolonized mice (left panel). Top correlated genes (Spearman rho>0.6) are highlighted in black and parsed for enrichment of biological pathways using Enrichr. Top pathways from GO Molecular Function, with corresponding adjusted p.values and gene names, are shown (right panel).
See also Table S5.
We then assessed the impact of bacterial exposure on AMP transcription in the intestines. The property of high variability in the GF colon was maintained upon microbial exposure (Fig. 6B). Expression of most AMPs was not substantially affected in the SI of any of the monocolonized mice, with only a modest induction of Reg3 family transcripts by a few bacterial species (Fig. 6C). The most profound change in the SI was a down-regulation of all three Reg3 genes by F. varium. In marked contrast, AMP expression was more responsive in the colon, with changes extending significantly beyond the baseline fluctuation in GF colons (Fig. 6D). Many α-defensin (but not β-defensin) transcripts were coordinately induced by a few phylogenetically diverse species (e.g., Parabacteroides merdae, Porphyromonas uenonis), with a similar pattern for the Reg3 family.
As denoted by the high CV of AMP transcripts in the colon (Fig. 6A), individual GF mice manifested substantial differences in the expression of α-defensin and Reg3 genes. This fluctuation in AMP levels, even in the absence of microbes, suggested that other triggers were affecting their expression. To elucidate the source of this variability, we searched for other genes whose expression correlated with AMPs across the colons of either GF or monocolonized mice (Fig. 6E, left panel). There was no correlation with the expression of IFN signature genes, which would have indicated enteric viral infections, or with Il22 transcripts, which would have suggested stimulation of epithelial cells by ILCs via Il22. A group of genes stood out as most strongly correlated with AMPs in both GF and colonized mice; pathway analysis of these transcripts revealed a significant enrichment in a number of nutrient transport and lipid metabolism pathways, suggesting a link among nutrition, enterocyte function, and AMP production (Fig. 6E, right panel; Table S5). Thus, colonization by some symbionts elicits highly coordinated AMP expression in the colon over a fluctuating background that appears to reflect intestinal function rather than microbial stimulation.
Fusobacterium variun elicits an unusually strong host response signature.
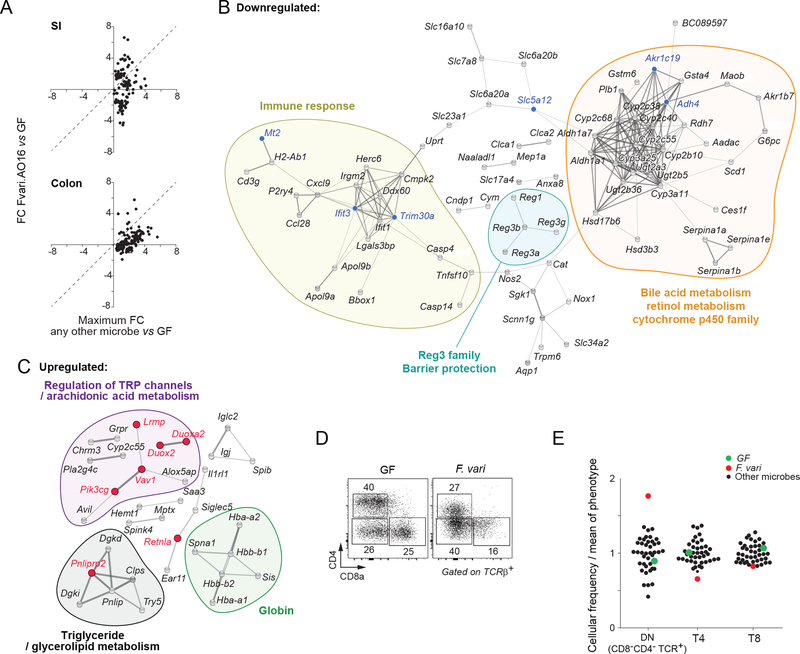
It was clear from the gene-expression data of Figs. 4 and 6 that F. varium was one of the more stimulatory bacteria. Interestingly, F.varium also influenced many immune cell populations in the colon (Fig. 2C, especially double-negative (DN) T cells). F. varium is a gram-negative obligate anaerobe in the phylum Fusobacteria. In the SI, monocolonization with this species stood out, with a concentrated suppression of genes within cluster 2 and a strong up-regulation of cluster 7 (Fig. 4C). In the colon, its effects were also strong, albeit less unusual (Fig. 4D).When the SI transcriptomes of mice colonized with F. varium (AO16) were compared with the transcriptomes of any other monocolonized mice, 35% of the genes were more strongly induced (Fig. 7A). Seven percent of this set of genes were also more intensely induced in the colon by F. varium than by other bacteria. (Fig. 7A).
Figure 7: Host response to Fusobacterium varium.
(A) Amplified gene expression preferential to F. varium (Fvari.AO16), based on the conservative gene list established in Fig. 4B–C. Fold change (FC) of Fvari.AO16 over GF (y-axis) was compared to the maximum induced FC by any other microbe over GF (x-axis). Top – SI, bottom- colon.
(B) Functional analysis of genes suppressed by F. varium.
STRING-db clustering and functional categories of significantly altered genes (FC≤0.5 in SI; FC≤0.67 in colon vs. GF; FDR 0.1). Blue dots - genes from (A) preferentially suppressed by Fvari.AO16; gray dots - all other suppressed genes in the Fvari.AO16 response that formed connected clusters. Functional categories determined by GO and KEGG are shown: “Retinol metabolism” FDR 2.25e-15. “Bile acid metabolism” FDR 2.6e-7. “Immune response” FDR 0.0138.
(C) Functional analysis of genes induced by F. varium.
STRING-db clustering and functional categories of significantly altered genes (SI FC≥2, colon FC≥1.5 vs. GF; FDR 0.1). Red dots – genes from (A) preferentially induced by Fvari.AO16-; gray dots - all other induced genes in Fvari.AO16 response that formed connected clusters. Functional categories determined by GO and KEGG: “Regulation of TRP channels” FDR 0.00313; “AA metabolism” FDR 0.0241; “Globin” FDR 3.78e-8; “Triglyceride metabolism” FDR 0.0184; “Glycerolipid metabolism” FDR 1.32e-7.
(D) Representative flow cytometry plots of CD4 and CD8 expression in GF and Fvari.AO16, gated on CD45+ CD19-TCRβ+ cells.
(E) Frequencies of T4, T8, and DN T cells normalized to the mean frequency of all microbes in all monocolonizations.
See also Tables S6A–B.
We investigated the functional nature of the response to F. varium by clustering (in the String database) the sets of transcripts down- or up-regulated by F. varium in either the SI or the colon (Figs. 7B and 7C). Overall, there were a few altered genes related to immune function. Repressed transcripts included a large set related to bile acid metabolism, with a sizable cluster of the Cytochrome p450 gene family (e.g., Cyp3a25, Cyp2b10) and retinol metabolism genes (e.g., Rdh7, Aldh1a1) (Fig. 7B). Cytochrome p450 controls mechanisms of xenobiotic metabolism in the gut (Carmody and Turnbaugh, 2014) and, together with other members of this cluster (e.g., Rdh7 or Aldh1), influences the metabolism of all _trans_-retinoic acid. F. varium also strongly repressed the Reg3 antimicrobial family, particularly in the SI (Fig. 6C). One can speculate that an advantage is gained by F. varium in suppressing these AMPs, which are thought to play an important role in barrier integrity and are usually induced by microbes (Wang et al., 2016; Vaishnava et al., 2008; Cash et al., 2006). Perhaps F. varium suppresses Reg3 to avoid death induced by AMPs, creating a more favorable mileu for itself. Up-regulated genes include those involved in arachidonic acid metabolism (e.g., Alox5ap) (Fig. 7C), the essential precursor for lipid mediators of inflammation.
In accordance with the transcriptional effects, F. varium had one of the largest phenotypic impacts (Fig. 2D). Specifically, it had the strongest effect on αβT cells, reducing both T4 (CD4+) and T8 (CD8+) populations and causing a higher frequency of colonic DN (CD4-CD8-TCRβ+) cells than any other microbe (Figs. 7D and 7E).
Fusobacterium spp. are among the few intestinal symbionts that can be found in both vertebrates and in free-living bacterial communities, rendering them potent to introduce evolutionarily honed functions (Ley et al., 2008). Relatively little is known about the Fusobacterium genus and human health, but Fusobacterium nucleatum is prevalent among patients with colorectal carcinoma and among some patients with inflammatory bowel disease (Kostic et al., 2013; Strauss et al., 2011). The virulence and invasiveness of F. nucleatum strains vary via unknown mechanisms that do not fit subspecies classifications (Strauss et al., 2011), and the strain of F. nucleatum used here (F0419) elicited no outstanding phenotypes in our study. However, F. varium’s prominent signature supports the notion that members of this genus may have unique interactions with the host.
DISCUSSION
The driving concept of this study was that the gut microbiota hosts a largely untapped wealth of immunomodulatory activities. To provide proof of concept, we devised a sensitive, broad-ranging screen that entailed monocolonization of mice with human gut symbionts followed by extensive, unsupervised immunophenotyping and transcriptomics. Indeed, a screen of 53 bacterial species yielded a number of activities, both anticipated and unanticipated. For example, we found individual microbes capable of inducing Th17 cells in the SI to a level similar to that driven by SFB, as is explored in depth elsewhere (Tan et al., 2016). Unexpected, was the observation that about one-quarter of the bacteria examined, encompassing a diversity of species, could induce RORγ+Helios- Tregs in the colon (Sefik et al., 2015), given claims that a consortium of 17 Clostridium species or several limited individual members of the microbiota are needed for Treg induction (Atarashi et al., 2013, Faith et al., 2014). Other potentially interesting immunomodulatory activities have not been reported previously—e.g., the augmentation of Il10-producing CD4+ T cells and the parallel reduction of Il22-producing ILCs in the colon by Veillonella; the impressive reduction of pDC numbers by L. rhamnosus; and the unusually strong and broad immunoperturbing activity of F. varium.
Thus, this approach has the potential to yield an apothecary of immunomodulatory agents tailored to modulate the immune system in a chosen manner. No doubt local gut effects will be the most straightforward to achieve, but precedents indicate that microbiota manipulations can regulate gut-distal immune responses—both protective and pathogenic (Wu et al., 2010; Ochoa-Reparaz et al., 2010; Hsiao et al., 2013). Future efforts will be devoted to evaluate selected immunomodulatory microbes in hosts with a substantially more complex intestinal microbiota than monocolonized mice, particularly humans. Data on RORγ+Helios- Tregs and Th17 cells argue that at least some of the observed activities can be recapitulated in SPF mice (Sefik et al., 2015; Tan et al., 2016). An alternative strategy would be to identify molecules and pathways underlying the immunomodulatory activity of interest. The value of this microbiota-based apothecary will increase in parallel with our molecular and cellular understanding of the immunologic diseases we aim to target, allowing a more precise choice of immunomodulatory activity. Lastly, it will be interesting to see whether extending the screen beyond the 53 bacterial species evaluated here will identify additional activities.
Beyond these practical considerations, our data provide several insights into immune system–microbiota interactions in the gut. The enormous complexity of the intestinal microbiota means that isolating the impact of a particular bacterial species on the intestinal or systemic immune system is a rather daunting task. Our reliance on gnotobiotic conditions aids such deconvolution. Importantly, we found that, in the absence of competition, most of the tested bacteria were able to robustly colonize the mouse intestine and that the great majority of them elicited immunophenotypic and/or transcriptomic changes, while few were stealth to the parameters measured. We had previously reported (Chung et al., 2012) that mice colonized with a complex human microbiota had small intestinal immune systems characteristic of GF mice. In contrast, here we show that colonization with single microbes derived from the human intestine does influence the immune system in the gut of host mice. We ascribe the different outcomes to the much higher load of any one bacterium (up to 10,000x higher in monocolonized mice than in “human microbiota” mice), providing much greater antigen or metabolite stimuli.
Another message conveyed by our data is that immune system recalibration to the microbiota shows substantial diversity and redundancy. On one hand, most microbes elicited a distinct immunophenotype in the host; on the other hand, many immunologic alterations were induced by more than one microbe, and bacteria could be found with opposite effects in most parameters. We speculate that these adaptations might explain why microbial communities are so vast, providing balance to both the community and the host. A sufficiently large community of diverse genomic inputs allows buffering in case certain community members are lost. The broad diversity and redundancy of immunologic alterations permit many different microbes to provide the balance needed to promote overall host health. Importantly, both the diversity and the redundancy can be provided by organisms from the same or different phyla. Similarly, none of the transcriptional effects were induced by all of the microbes. In fact, different bacteria often had opposing impacts on the gut transcriptome, for example AMP gene expression. There did not appear to be a phylogenetic relationship in either the immunologic or the genomic changes. The lack of a relation between microbe-induced immune recalibration and microbial phylogeny would also contribute to stabilization of the microbiota’s influence even if specific taxa were lost. The bacteria examined induced both shared and unique responses in different tissues at both the transcriptional and the cellular levels. For example, for Tregs and pDCs, a strong correlation existed between the SI and the colon (and other tissues). However, for Il17, Il22, and ILCs, recalibration and transcriptional responses to bacteria were mostly restricted to the SI. Interestingly, the finding of greater variability between gene-expression profiles in GF mice than in monocolonized mice supports the contention (Quackenbush, 2002) that the presence of microbial communities stabilizes both immunologic and transcriptional phenotypes and provides resistance to perturbation. This notion of coupled diversity and redundancy may also explain why it is so often difficult to distill a designated microbiota influence or state of dysbiosis down to a single (or a single set of) bacterial species.
The absence of outcomes shared by all species within a phylum, or even a genus, suggests that this interspecies diversification might have occurred through horizontal transfer and/or that the corresponding mechanisms/pathways are common in the bacterial world. Moreover, our study shows differences even among the strains of the same species. This highlights the importance of strain specificity being associated with immunophenotypes. Even in parallel colonizations with the same microbes, we observed some differences. It is certainly possible that the bacterial and host transcriptomes adapt at different rates and that factors other than the ones we controlled for, such as microbial load, host age, and duration of colonization, are important in stabilizing responses.
This study is remarkable in demonstrating that the gut microbiota presents many opportunities to impact the host immune system. It is clear that multiple individual microbes have important effects on the host, and that a balance of the microbiota is necessary for homeostasis. It would be interesting to study the combinatorial effects of immunomodulatory microbes both in a gnotobiotic setting and also under SPF conditions. Determining the minimal consortium of microbes that can maintain a stable balance between the microbiota and the host immune system will likely now be possible. By having identified individual effector strains, future studies on the mechanisms of host/microbial interactions (pathway interactions and key molecules) are vital questions to be answered. Can one rationally control the microbiota to prevent and/or cure disease states, by treating with certain desired microbes or molecules? The advantage of using specific molecules which can be dosed and regulated as any drug, would yield host responses that are more reproducible and therefore advantageous over using viable bacteria to modify or regulate a given host response.
STAR METHODS AND RESOURCES
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Prof. Dennis Kasper dennis_kasper@hms.harvard.edu.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacteria
Bacteria were purchased obtained from several sources: the ATCC (atcc.org), BEI, (beiresources.org), or DSMZ (dsmz.de) repository or were obtained from BWH clinical labs or Harvard-affiliated labs (Table S1). Anaerobic bacteria were cultured PYG broth under strictly anaerobic conditions (80% N2, 10% H2, 10% CO2) at 37°C in an anaerobic chamber. All bacteria (Bacteroides, Clostridium, Bifidobacterium, Lactobacillus, Enterococcus, Fusobacterium, Propionibacterium, and Peptostreptococcus spp.) were grown in peptone—yeast––glucose medium supplemented with hemin and vitamin K or on brucella blood agar plates and TSA blood agar plates (BBL). Acinetobacter spp. were grown in Super Broth (SB) medium and on LB agar plates. Lachnospiraceae, Veillonella spp., and Coprobacillus spp. were grown in chopped meat broth. Staphylococcus spp. were grown aerobically at 37°C in L-broth and on LB agar plates. Campylobacter and Helicobacter spp. were grown on brucella blood agar plates (VWR) and kept in microaerophilic conditions (CampyPak EZ in an anaerobic container system) at 37°C. The cladogram was generated using Human Microbiome Project data in GraPhlAn (http://huttenhower.sph.harvard.edu/galaxy/) and MetaPhlAn version 1.1.0 (http://www.hmpdacc.org/HMSMCP/healthy/#data). The overall mean diversity calculated by MEGA6 was 0.472. The total mean abundance was 62.6 and the prevalence ranged from 1.4 to 100 with a median of 64.4.
All strains of bacteria that were not from international repositories (Table S1A) were deposited to BEI resources (https://www.beiresources.org/).
Mice
GF C57BL/6J mice were originally purchased from the National Gnotobiotic Rodent Resource Center of the University of North Carolina at Chapel Hill, and bred in our lab facility were used at Harvard Medical School in GF flexible film isolators (Class Biologically Clean®) throughout this study. Sterility tests (culture and PCR) were done every week, ensuring that mice remained GF. Mice food was autoclaved at 128°C for 30 min at 26 PSI. Water was autoclaved at 121°C for 1 h. SPF mice were housed under the same conditions in the same facility with the same food (autoclaved to ensure comparable nutrients) for 2 weeks. Animals of both genders were used as available. Littermates were randomly assigned to experimental groups, to avoid any bias, whenever possible. Animal protocol IS00000187 and COMS protocol 07–267 were approved by Harvard Medical School’s Institutional Animal Care and Use Committee and the Committee on Microbiological Safety, respectively. This study adheres to the ARRIVE guidelines.
Generation and processing of monocolonized mice
GF C57BL/6 mice were orally inoculated by gavage with a broth grown single bacterial strain at 4 weeks of age and kept in gnobiotic isolators. Each group of mice was housed in gnobiotic isolators under sterile conditions for 2 weeks. Fecal material was collected and plated at 1 week and 2 weeks after bacterial inoculation to ensure monocolonization by a single bacterial strain. The identity of all colonizing microbial species was confirmed by 16S sequencing using the 27F and 1492R primers and Sanger sequencing at the Harvard Biopolymers Facility. All colonizations were done and processed at the same time of the day to reduce diurnal variability. Processing was undertaken by the same individuals throughout these studies to minimize person-to-person variability.
METHOD DETAILS
Preparation of lymphocytes and flow cytometry
Intestinal tissues were treated with 30 mL of RPMI containing 1 mM dithiothreitol, 20 mM EDTA, and 2% FBS at 37°C for 15 min to remove epithelial cells. The intestinal tissues and Peyer’s patches were then minced and dissociated in RPMI containing collagenase II (1.5 mg/mL; Gibco), dispase (0.5 mg/mL), and 1% FBS, with constant stirring at 37°C (45 min for colons and small intestines; 15 min for Peyer’s patches). Single-cell suspensions were then filtered and washed with 4% RPMI solution.
Mesenteric lymph nodes (mLN), and Systemic lymphoid organs (SLO) were mechanically disrupted. Subcutaneous (inguinal and axillary) lymph nodes and spleens were pooled and red blood cells were lysed. To minimize variability and reagent drift, collagenase II and dispase were purchased in bulk and tested for consistency in digestion and viability of cells before use. Single-cell suspensions were stained for surface and intracellular markers and analyzed with BD LSRII.
Single-cell suspensions were stained with three constant panels of antibodies for consistency. The first panel included antibodies against CD4, CD8, TCRß, CD45, TCRγδ, CD19, Foxp3, Helios and Rorγ. The second panel included antibodies against CD45, CD4, TCRß, TCRγδ, Il17a, IFNγ, Il22, and Il10. The third panel included antibodies against CD45, CD19, CD11c, CD11b, Ly6c, PDCA-1, F4/80, and CD103. For cytokine analysis (second antibody panel), cells were treated with RMPI containing 10% FBS, phorbol 12-myristate 13-acetate (10 ng/mL; Sigma), and ionomycin (1 μM; Sigma) in the presence of GolgiStop (BD Biosciences) at 37°C for 3.5 h. For intracellular staining of cytokines and transcription factors (first and second antibody panels), cells were stained for surface markers and fixed in eBioscience Fix/Perm buffer overnight, with subsequent permeabilization in eBioscience permeabilization buffer at room temperature for 45 min in the presence of antibodies. Cells stained with the third panel of markers were fixed in 1% formalin diluted in DMEM overnight. Great care was taken to reduce variability and reagent drift in all enzymes, reagents and antibodies. Cells were acquired with a BD LSRII, and analysis was performed with FlowJo (Tree Star) software.
Compensation for each experiment was adjusted with Rainbow Calibration particles to ensure consistency in data collection. The concentration, clone, and source of antibodies were kept constant to ensure consistency in staining. Occasionally, the entire set of data was sampled and reanalyzed blindly to ensure equal gating criteria and scoring. The raw data were independently analyzed by two individuals, and an average value was reported. Each analyst used the same version of FlowJo Software and the same bio-exponential settings previously determined for each experiment. In rare cases of disagreement (i.e., when independent scoring differed by ≥25%), the scoring was re-determined by the two analysts together in order to understand and resolve the variation. If the disagreement could not be resolved, the data were excluded from the final reports. Any strong discrepancies in staining due to reagent drift (e.g., enzymes, antibodies) were noted, and the data in question were excluded from the final reports. Frequencies of each cell type were averaged for each microbial colonization condition.
IgA ELISA
IgA levels in feces of monocolonized mice were measured with a Mouse IgA Elisa Kit (eBioscience, 88–50450-88) according to the manufacturer’s instructions.
Gene-expression profiling
Data collection:
The same segments of the distal colon and (0.5 cm long and 3 cm away from rectum) and three segments (each 0.3cm long) from the same midsection of the duodenum, jejunum, and ileum of the small intestine were collected from mice. These segments were then homogenized in TRIzol and stored at −80°C until RNA isolation. GF samples were collected throughout the duration of the screen. Samples were collected from both female or and male mice. Colon profiling included a total of four batches of samples totaling in 56 samples from male mice and 16 samples from female mice. SI profiling included a total of four batches of samples totaling in 51 samples from male mice and 7 samples from female mice. Each batch of microbially colonized intestines was profiled together with at least two replicates of GF control samples. Profiling was performed on Affymetrix Mouse Genome M1.0 ST arrays as previously described (Cipolletta et al., 2012), nearly always at least in duplicates (singletons in rare instances).
QUANTIFICATION AND STATISTICAL ANALYSIS
Immunophenotypes
Fold-change values were calculated by dividing the frequencies of a given cell type for each microbial colonization by the average frequency obtained from GF mice, To control for multiple testing, a false discovery rate was calculated by the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995)was calculated and; the thresholds used are indicated in the text and figures where relevant.
Pearson correlations (for normalized mean immunophenotypes) and Euclidean distances (either per mouse or per normalized mean) within phyla, genera, species or strains were calculated by GeneE. To normalize per cell type, each frequency was divided by the mean of the cell type of interest across all microbes.
Gene expression profiling:
Data normalization and batch correction:
Microarray data were background-corrected and normalized with the robust multi-array average algorithm. Gender and batch effects were corrected in a linear model with the feature as dependent variable and technical variables (batches) as regressors (implemented by R package “swamp”).
CV calculation:
Microarrays for each microbe were typically performed in duplicate or triplicate. Thus, the CV per transcript for GF intestines was determined by (1) calculating the CV per transcript for randomly sampled GF pairs from a total of 8 (SI) or 12 (colon) GF replicates, and (2) iterating the random sampling 250 times and taking the average of the 250 CV values as the final CV value for GF mice. CV values for microbially colonized samples were calculated as per normal, without random sampling.
Selection of differentially expressed genes:
Analysis on the whole tissue transcriptome focused on a select set of genes with a fold change relative to GF of >2.5 (or <0.4) and uncorrected p(-log10)>2.5. Scatter analysis for most extreme effects on transcripts (both as fold change and as t-test p.value) was performed in R-Project or Multiplot Studio.
AMP aggregate score and correlation with gene expression:
Aggregate AMP scores were calculated as follows: (1) RNA levels for each transcript belonging to the α-defensin and Reg3 family of AMPs, for which changes in expression levels were most dynamic, were normalized to the mean expression level across all samples; and (2) the normalized transcript levels were then summed and averaged for each sample to derive an aggregate AMP score. The correlation of all other transcripts with the respective AMP scores was determined with the Spearman correlation test. Correlations were calculated separately for GF and colonized mice, with use of six randomly sampled replicates for either group and iteration of the sampling and correlation test 50 times. The mean of the 50 correlation coefficients was taken to be the final coefficient value. RNAs with a correlation coefficient of >0.6 for both GF and monocolonized mice were extracted for pathway enrichment analysis.
Clustering and enrichment analysis:
Hierarchical clustering and K-means clustering were performed on these selected genes in GeneE. Pathway analysis was done with STRING (www.string-db.org), and Enrichr (Chen et al., 2013; Kuleshov et al., 2016, http://amp.pharm.mssm.edu/Enrichr/). Enrichment for cell types was verified in ImmGen and GNF databases.
DATA AND SOFTWARE AVAILABILITY
The extensive dataset presented in Fig. 1–4, are available for mining (Tables S1, S2, S3). Phylogenetic identity of all bacteria is detailed in Table S1A. The immunophenotypes as frequencies of cell types per an invidiual mouse basis are presented in Table S2D. The gene expression raw data are available in the GEO with accession number GSE88919.
Supplementary Material
Figure S1A-S1C
Figure S1, Related to Fig. 1: Flow cytometry gating strategies. Representative flow cytometry plots demonstrating the gating strategy for the three staining panels: lymphocytes (A), myeloid cells (B), and the cytokines (C).
Figure S2A-S2F
Figure S2, Related to Fig. 2, Table S2: Immune cell populations and IgA induction by the gut bacteria
(A) Rank-ordered average frequencies of each immunocyte population for every monocolonized microbe in SI and PP as measured by flow cytometry.
(B)Rank-ordered average frequencies of each immunocyte population for every monocolonized microbe in mLN and SLO, as measured by flow cytometry.
For cell-type frequency determination (y-axis) and bacterial identification (x-axis), see Tables S1B, S2A, and S2B. For gating strategies, see Fig. S1.
(C) Representative flow cytometry plots of monocytes (Ly6c+CD11b+) in the SI (gated on CD45+CD19- cells). Monocytes include Ly6chi and Ly6clo populations, which are measured as a uniform population in the quantification. Plots here highlight that certain microbes can induce Ly6chi, Ly6clo, or both.
(D) Representative flow cytometry plots of CD11b and CD11c expression in the SLO (gated on CD45+CD19- cells). These populations correspond to macrophages, F4/80+ mononuclear phagocytes, CD103+ DCs, and pDCs. CD11b expression is dimmer in the SLO compared to intestinal tissues. The CD11bloCD11clo population, which is largely absent in the intestines, is more pronounced in the SLO. These qualities of myeloid cells were not reflected in the quantification in Figs. 2A and 2B.
(E) Representative flow cytometry plots of T4, T8 and DN T cells (gated on CD45+TCR+CD19- cells) in the SI. In contrast to the majority of myeloid markers, the lymphocyte markers are clearer and more consistent across tissues.
(F) Fecal IgA induction of individual monocolonized mice. IgA concentration quantified by ELISA (upper), %IgA quantified by flow cytometry (lower).
Figure S3A-S3B
Figure S3, Related to Fig. 3: Correlations of immunophenotypes across tissues. (A) Pearson correlations were performed for each cell population assayed in the SI, colon, mLN, and SLO, and the resulting correlation coefficients were plotted as a heat map. Three correlated clusters were evident: CD11b+F4/80+ cells (which encompass CD11b+CD11c- MF and CD11b+CD11c+ MNPs), monocytes, Foxp3-RORγ+CD4+ T cells (as a proxy for T4 cells capable of Il17 production), and a Foxp3+RORγ+Helios- Treg cluster (measured separately as Foxp3+Helios- or RORγ+Helios-). (B) Pearson correlation of the overall immunologic impact of microbes on the SI and colon. Values for each immunophenotype were normalized to the mean across all microbes. Hierarchical clustering was performed.
Figure S5
Figure S5, Related to Fig. 5: Colonic DCs and pDCs frequencies in monocolonized mice. Frequencies of CD103+CD11b- DCs (top; gated on CD45+CD19- cells) and of pDCs (bottom; gated on CD45+CD19-CD11b- cells) induced in the colon by monocolonizing microbes. Microbes were ordered according to their pDC induction level and color-coded for individual experiments. GF data are shown in green.
Figure S4A-S4D
Figure S4, Related to Fig. 4: The host transcriptional responses to monocolonization. (A) Volcano plot [p(-log10) vs. Fold Change] representations of the microarray data in the colon. (B) Volcano plot [p(-log10) vs. Fold Change] representations of the microarray data in the SI. (C) Levels of Il18 transcript across the microbes studied in the colon. (D) Levels of Il18 transcript across the microbes studied in the SI.
Table S1A-S1C
Table S1, Related to Fig. 1: Microbial strains/colonization, and cell types
(A) Complete list of microbes in this study. “Microbe_Name” includes the species name and the strain identification; “Key_Microbe_Name” and “Abbreviation” indicate short versions of the Microbe_Name used throughout the paper. “Origin” specifies the source from which the microbe can be obtained. The 16S NCBI match is provided for bacterial species that did not match their original classification.
(B) Cell specifications of cell types, their markers, and gating strategies.
(C) Colony-forming units in feces (per gram) in the mLN and SLO for all microbes in this study.
Table S3
Table S3, Related to Fig. 4: Fold Change of colonic and small-intestinal transcripts that are most impacted by monocolonization.
Table S4
Table S4, Related to Fig. 5: List of genes reproducibly correlated to pDC frequency in both SI and colon. Correlation coefficients are shown.
Table S6A-S6B
Table S6, Related to Fig. 7: Transcriptional response to F. varium colonization
(A) Complete list of genes that are up- or down-regulated in the SI and colon of _Fusobacterium varium_–colonized mice. FC (Fvari.AO16/GF) ≤0.5 (repressed) and ≥2 (induced) for SI and FC Fvari.AO16/GF) ≤0.67 (repressed) and ≥1.5 (induced) for colon.
(B) List of _F. varium_–preferential genes. These genes are most strongly altered in _F. varium_–colonized mice compared with mice colonized with any other microbe [FC (varium.AO16/other microbes) cut off 1.5].
Table S5
Table S5, Related to Fig. 6: Gene correlations with AMP scores in GF and monocolonized mice. Spearman correlation coefficients are shown.
Table S2A-S2D
Table S2, Related to Fig. 2: Complete immunologic dataset
(A) Mean frequencies of all cell types for each microbe ± standard deviations.
(B) Fold-Change values (compared to GF) for each microbe.
(C) Qualitative phenotypic changes in immune cells.
(D) Frequencies of all cell types for each microbe per mouse, including all strains of microbes in this study.
ACKNOWLEDGMENTS
We thank Drs. A. Onderdonk, L. Bry, A. DuBois, M. Delaney, H. Renz, L. Comstock, J. Mekalanos, A. Sirota-Madi, Y. Benita, A. Madi and N. Surana for mice, microbial strains, or insightful discussion, J. Ramos, S. Edwards, A. Rhoads, H. Paik and C. Laplace for help with mice, profiling and figure designs. Some microbes were obtained through BEI Resources, the NIAID, and the NIH as part of the Human Microbiome Project. Some microbes were from ATCC. Others were obtained from the HDDC center at Harvard - we thank the “Massachusetts Host Microbiome Center” and P30 Ctr grant from NIDDK. This work was funded by a Sponsored Research Agreement from UCB. N.G.-Z. was supported by HFSP (LT00079/2012) and EMBO (ALTF 251-2011) fellowships, a Fulbright Award, a UNESCO L’Oreal National and International Women in Science Award, and the Weizmann Institute of Science–Revson National Postdoctoral Award Program for Advancing Women in Science; E.S. by a fellowship from the Boehringer Ingelheim Fonds; L.K. by the National Science Foundation; and T.G.T. by an A*STAR Graduate Scholarship fellowship.
REFERENCES
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, and Kasper DL (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 156, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 500, 232–236. [DOI] [PubMed] [Google Scholar]
- Benjamini Y and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Roy. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bevins CL and Salzman NH (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9, 356–368. [DOI] [PubMed] [Google Scholar]
- Carmody RN and Turnbaugh PJ (2014). Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin. Invest 124, 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, and Hooper LV (2006). Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 313, 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Kaliaperumal N, Chretien AS, Tang S, Lee B, Poidinger M, Fairhurst AM, and Connolly JE (2014). Human plasmacytoid dendritic cells regulate IFN-alpha production through activation-induced splicing of IL-18Ralpha. J Leukoc. Biol 96, 1037–1046. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC. Bioinformatics. 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 149, 1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, and Mathis D (2012). PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 486, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Erturk-Hasdemir D, Ochoa-Reparaz J, Reinecker HC, and Kasper DL (2014). Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host. Microbe 15, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, and Littman DR (2013). Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 494, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, and Gordon JI (2014). Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl. Med 6, 220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara D, Wei B, Presley LL, Brewer S, McPherson M, Lewinski MA, Borneman J, and Braun J (2008). Systemic control of plasmacytoid dendritic cells by CD8+ T cells and commensal microbiota. J Immunol 180, 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De PM, Brandi G et al. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 31, 677–689. [DOI] [PubMed] [Google Scholar]
- Gallo RL and Hooper LV (2012). Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol 12, 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van TW, Ren B, Schwager E, Knights D, Song SJ, Yassour M et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host. Microbe. 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez de AM, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science. 351, 1296–1302. [DOI] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM et al. (2013). Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 498, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions between the microbiota and the immune system. Science. 336, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV and Macpherson AJ (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev. Immunol 10, 159–169. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 155, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, and Nunez G (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13, 321–335. [DOI] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, and Gordon JI (2011). Human nutrition, the gut microbiome and the immune system. Nature. 474, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop KA, McDonald KG, McCrate S, McDole JR, and Newberry RD (2015). Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal. Immunol 8, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole A, He J, Rivollier A, Silveira DD, Kitamura K, Maloy KJ, and Kelsall BL (2013). Type I IFNs regulate effector and regulatory T cell accumulation and anti-inflammatory cytokine production during T cell-mediated colitis. J Immunol 191, 2771–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen AM, Peet A, Tillmann V, Pöhö P, Mattila I, Lähdesmäki H, Franzosa EA, Vaarala O, de Goffau M, Harmsen H, Ilonen J, Virtanen SM, Clish CB, Orešič M, Huttenhower C, Knip M; DIABIMMUNE Study Group., Xavier RJ. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 17(2):260–73. (PMID:25662751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, and Hsieh CS (2011). Peripheral education of the immune system by colonic commensal microbiota. Nature. 478, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD et al. (2011). AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 13, 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, and Gordon JI (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol 6, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ and Uhr T (2004). Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 303, 1662–1665. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, and Kasper DL (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 122, 107–118. [DOI] [PubMed] [Google Scholar]
- Metidji A, Rieder SA, Glass DD, Cremer I, Punkosdy GA, and Shevach EM (2015). IFN-alpha/beta receptor signaling promotes regulatory T cell development and function under stress conditions. J Immunol 194, 4265–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, and Mathis D (2014). Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc Natl Acad Sci U S A. 111, 6696–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahashi-Oda C, Udayanga KG, Nakamura Y, Nakazawa Y, Totsuka N, Miki H, Iino S, Tahara-Hanaoka S, Honda S, Shibuya K et al. (2016). Apoptotic epithelial cells control the abundance of Treg cells at barrier surfaces. Nat Immunol 17, 441–450. [DOI] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, and Kasper LH (2010). A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal. Immunol 3, 487–495. [DOI] [PubMed] [Google Scholar]
- Quackenbush J (2002). Microarray data normalization and transformation. Nat. Genet 32 Suppl, 496–501. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Foster KR, and Comstock LE (2016). The evolution of cooperation within the gut microbiota. Nature. 533, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B, Bunin A, Ghosh HS, Lewis KL, and Sisirak V (2011). Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev Immunol 29, 163–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders A, Yessaad N, Vu Manh TP, Dalod M, Fenis A, Aubry C, Nikitas G, Escaliere B, Renauld JC, Dussurget O et al. (2011). Identity, regulation and in vivo function of gut NKp46+RORgammat+ and NKp46+RORgammat- lymphoid cells. EMBO J 30, 2934–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O et al. (2008). Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29, 958–970. [DOI] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, and Eberl G (2011). RORgt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol 12, 320–326. [DOI] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A et al. (2016). Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J et al. (2015). Individual intestinal symbionts induce a distinct population of RORg+ regulatory T cells. Science. 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PA (2015). The tantalizing links between gut microbes and the brain. Nature. 526, 312–314. [DOI] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, and Allen-Vercoe E (2011). Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel. Dis 17, 1971–1978. [DOI] [PubMed] [Google Scholar]
- Surana NK and Kasper DL (2014). Deciphering the tete-a-tete between the microbiota and the immune system. J Clin. Invest 124, 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M and Colonna M (2015). The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TG, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Kasper DL, Benoist C et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A. (2016). Proc Natl Acad Sci U S A.113, E8141–E8150. Epub 2016 Nov 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, and Hooper LV (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 105, 20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fouts DE, Starkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho SB, Brenner DA et al. (2016). Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe 19, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, and Mathis D (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 32, 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1A-S1C
Figure S1, Related to Fig. 1: Flow cytometry gating strategies. Representative flow cytometry plots demonstrating the gating strategy for the three staining panels: lymphocytes (A), myeloid cells (B), and the cytokines (C).
Figure S2A-S2F
Figure S2, Related to Fig. 2, Table S2: Immune cell populations and IgA induction by the gut bacteria
(A) Rank-ordered average frequencies of each immunocyte population for every monocolonized microbe in SI and PP as measured by flow cytometry.
(B)Rank-ordered average frequencies of each immunocyte population for every monocolonized microbe in mLN and SLO, as measured by flow cytometry.
For cell-type frequency determination (y-axis) and bacterial identification (x-axis), see Tables S1B, S2A, and S2B. For gating strategies, see Fig. S1.
(C) Representative flow cytometry plots of monocytes (Ly6c+CD11b+) in the SI (gated on CD45+CD19- cells). Monocytes include Ly6chi and Ly6clo populations, which are measured as a uniform population in the quantification. Plots here highlight that certain microbes can induce Ly6chi, Ly6clo, or both.
(D) Representative flow cytometry plots of CD11b and CD11c expression in the SLO (gated on CD45+CD19- cells). These populations correspond to macrophages, F4/80+ mononuclear phagocytes, CD103+ DCs, and pDCs. CD11b expression is dimmer in the SLO compared to intestinal tissues. The CD11bloCD11clo population, which is largely absent in the intestines, is more pronounced in the SLO. These qualities of myeloid cells were not reflected in the quantification in Figs. 2A and 2B.
(E) Representative flow cytometry plots of T4, T8 and DN T cells (gated on CD45+TCR+CD19- cells) in the SI. In contrast to the majority of myeloid markers, the lymphocyte markers are clearer and more consistent across tissues.
(F) Fecal IgA induction of individual monocolonized mice. IgA concentration quantified by ELISA (upper), %IgA quantified by flow cytometry (lower).
Figure S3A-S3B
Figure S3, Related to Fig. 3: Correlations of immunophenotypes across tissues. (A) Pearson correlations were performed for each cell population assayed in the SI, colon, mLN, and SLO, and the resulting correlation coefficients were plotted as a heat map. Three correlated clusters were evident: CD11b+F4/80+ cells (which encompass CD11b+CD11c- MF and CD11b+CD11c+ MNPs), monocytes, Foxp3-RORγ+CD4+ T cells (as a proxy for T4 cells capable of Il17 production), and a Foxp3+RORγ+Helios- Treg cluster (measured separately as Foxp3+Helios- or RORγ+Helios-). (B) Pearson correlation of the overall immunologic impact of microbes on the SI and colon. Values for each immunophenotype were normalized to the mean across all microbes. Hierarchical clustering was performed.
Figure S5
Figure S5, Related to Fig. 5: Colonic DCs and pDCs frequencies in monocolonized mice. Frequencies of CD103+CD11b- DCs (top; gated on CD45+CD19- cells) and of pDCs (bottom; gated on CD45+CD19-CD11b- cells) induced in the colon by monocolonizing microbes. Microbes were ordered according to their pDC induction level and color-coded for individual experiments. GF data are shown in green.
Figure S4A-S4D
Figure S4, Related to Fig. 4: The host transcriptional responses to monocolonization. (A) Volcano plot [p(-log10) vs. Fold Change] representations of the microarray data in the colon. (B) Volcano plot [p(-log10) vs. Fold Change] representations of the microarray data in the SI. (C) Levels of Il18 transcript across the microbes studied in the colon. (D) Levels of Il18 transcript across the microbes studied in the SI.
Table S1A-S1C
Table S1, Related to Fig. 1: Microbial strains/colonization, and cell types
(A) Complete list of microbes in this study. “Microbe_Name” includes the species name and the strain identification; “Key_Microbe_Name” and “Abbreviation” indicate short versions of the Microbe_Name used throughout the paper. “Origin” specifies the source from which the microbe can be obtained. The 16S NCBI match is provided for bacterial species that did not match their original classification.
(B) Cell specifications of cell types, their markers, and gating strategies.
(C) Colony-forming units in feces (per gram) in the mLN and SLO for all microbes in this study.
Table S3
Table S3, Related to Fig. 4: Fold Change of colonic and small-intestinal transcripts that are most impacted by monocolonization.
Table S4
Table S4, Related to Fig. 5: List of genes reproducibly correlated to pDC frequency in both SI and colon. Correlation coefficients are shown.
Table S6A-S6B
Table S6, Related to Fig. 7: Transcriptional response to F. varium colonization
(A) Complete list of genes that are up- or down-regulated in the SI and colon of _Fusobacterium varium_–colonized mice. FC (Fvari.AO16/GF) ≤0.5 (repressed) and ≥2 (induced) for SI and FC Fvari.AO16/GF) ≤0.67 (repressed) and ≥1.5 (induced) for colon.
(B) List of _F. varium_–preferential genes. These genes are most strongly altered in _F. varium_–colonized mice compared with mice colonized with any other microbe [FC (varium.AO16/other microbes) cut off 1.5].
Table S5
Table S5, Related to Fig. 6: Gene correlations with AMP scores in GF and monocolonized mice. Spearman correlation coefficients are shown.
Table S2A-S2D
Table S2, Related to Fig. 2: Complete immunologic dataset
(A) Mean frequencies of all cell types for each microbe ± standard deviations.
(B) Fold-Change values (compared to GF) for each microbe.
(C) Qualitative phenotypic changes in immune cells.
(D) Frequencies of all cell types for each microbe per mouse, including all strains of microbes in this study.
Data Availability Statement
The extensive dataset presented in Fig. 1–4, are available for mining (Tables S1, S2, S3). Phylogenetic identity of all bacteria is detailed in Table S1A. The immunophenotypes as frequencies of cell types per an invidiual mouse basis are presented in Table S2D. The gene expression raw data are available in the GEO with accession number GSE88919.