ComplexContact: a web server for inter-protein contact prediction using deep learning (original) (raw)
Abstract
ComplexContact (http://raptorx2.uchicago.edu/ComplexContact/) is a web server for sequence-based interfacial residue-residue contact prediction of a putative protein complex. Interfacial residue-residue contacts are critical for understanding how proteins form complex and interact at residue level. When receiving a pair of protein sequences, ComplexContact first searches for their sequence homologs and builds two paired multiple sequence alignments (MSA), then it applies co-evolution analysis and a CASP-winning deep learning (DL) method to predict interfacial contacts from paired MSAs and visualizes the prediction as an image. The DL method was originally developed for intra-protein contact prediction and performed the best in CASP12. Our large-scale experimental test further shows that ComplexContact greatly outperforms pure co-evolution methods for inter-protein contact prediction, regardless of the species.
INTRODUCTION
Most proteins function by interacting with others to form complexes and/or protein-protein interaction (PPI) networks (1). Solving the structures of protein complexes by experimental techniques is very challenging (2). For example, there is little structural information for ∼80% of currently known protein interactions in bacteria, yeast or human (3). Computational prediction is an alternative way to elucidate the structure of a complex of interacting proteins. Inter-protein contact prediction is becoming an important intermediate step for such a task (4,5).
Due to the evolution pressure, co-evolved residues are often found to be spatially proximal within the protein structure (6) or upon the protein-protein interface (7). As such, co-evolution analysis or more specifically direct-coupling analysis (DCA) (e.g. EVcomplex (5) and Gremlin-Complex (4)) is widely used to identify co-evolved residues and predict inter-residue contacts from multiple sequence alignments (MSA) (6–15). Although popular, DCA has low accuracy when a protein under prediction does not have many sequence homologs in MSA (16–23). This problem becomes even more serious for inter-protein contact prediction since it is challenging to find so many interlogs (i.e. interacting homologs) for an interacting protein pair, especially for eukaryotic species (4,5).
We present ComplexContact, a web server that predicts inter-protein residue-residue contacts without using any structural templates. The underlying algorithm of this server is a Deep Learning (DL) model which has won intra-protein contact prediction in CASP12 (24,25). In addition to co-evolution information, our DL method makes use of sequential features and contact occurrence patterns to dramatically reduce the requirement of sequence homologs and greatly improve accuracy (16). This server also applies a phylogeny-based method to identify interlogs and build better MSAs for a protein pair from eukaryotes. Our experimental results show that ComplexContact outperforms pure DCA for inter-protein contact prediction for both prokaryotes and eukaryotes.
MATERIALS AND METHODS
The detailed description of the DL algorithm underlying ComplexContact is described in (16) and the detailed experimental results for inter-protein contact prediction is available at (26). Here, we briefly summarize the method.
Overall flowchart for inter-protein contact prediction
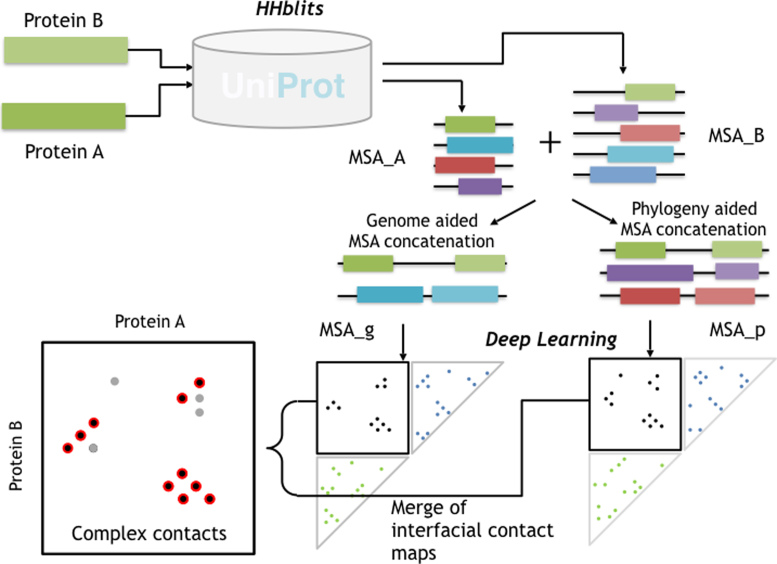
As shown in Figure 1, given a pair of putative interacting protein sequences A and B for which users would like to predict inter-protein contacts, our method first employs HHblits to search for sequence homologs and build MSAs for A and B, respectively. Then we employ two strategies (i.e. genome- and phylogeny-based) to concatenate MSA_A and MSA_B into two paired MSAs consisting of only interlogs, denoted as MSA_g and MSA_p, from which we may predict two inter-protein contact maps using our DL model (trained from single-chain proteins), and calculate their average as the final prediction.
Figure 1.
Illustration of ComplexContact workflow. Given a pair of putative interacting proteins A and B, ComplexContact first uses HHblits (38) to search for sequence homologs and build an MSA for each protein. Then ComplexContact constructs two paired MSAs using genome and phylogeny information. Finally, ComplexContact applies deep learning to predict two inter-protein contact maps from the two paired MSAs and calculates their average as the final contact prediction. The top half of this figure is inspired by Fig. S2 in (40).
Concatenate two multiple sequence alignments
Concatenating MSAs by genomic distance
In prokaryotes and some eukaryotes, interacting genes are often co-located on the chromosome into operons (27), so we may assume two proteins forming an interacting pair if their intergenic distance is less than a threshold (28). A similar approach is used in EVcomplex (5) and Gremlin-Complex (4).
Concatenating MSAs by phylogeny
In most eukaryotes, it is challenging to concatenate two individual MSAs since an individual MSA may contain abundant paralogs and two genes may interact even if they are not close by genomic distance (13). Here, we group proteins in each MSA by their species (or sub-species if possible) according to the phylogeny tree in the Taxonomy Database (29). Then we sort proteins of a specific species/subspecies in each MSA by their sequence similarity (from high to low) to their respective query proteins. Let p1, p2, …, p_m_ and q1, q2, …, q_n_ be the sorted proteins of a specific species in two MSAs, respectively. Then we pair p_i_ and q_i_ together where i ranges from 1 to the minimum of m and n.
On average for eukaryotes, the phylogeny-based method works better while for prokaryotes, the genomic-based method works better. Combining them can improve performance on eukaryotes. For some protein pairs, neither method can identify many sequence homologs, and the resultant interfacial contacts may have low accuracy.
Deep learning for inter-protein contact prediction
Our DL model is formed mainly by two deep residual neural networks (ResNet) (30). One is used to handle sequential features and the other pairwise features. The first ResNet conducts 1-dimensional (1D) convolutional transformation of sequential features to capture long-range sequential context of each residue in the query proteins. Its output is converted to a 2-dimensional (2D) matrix and then fed into the 2nd ResNet together with the original pairwise features. The second ResNet conducts 2D convolutional transformation of its input to capture long-range 2D context of a residue pair. Finally, the output of the 2nd ResNet is fed into logistic regression, which predicts the probability of any two residues forming a contact.
The sequential features include protein sequence profile, predicted 3-state secondary structure (31) and 3-state solvent accessibility (32,33). The pairwise features include the direct information produced by CCMpred (11) and mutual information derived from paired MSAs.
The DL model underlying ComplexContact was originally developed for intra-protein contact prediction, which has been officially ranked No. 1 in CASP12 (24,25). That is, our DL model was trained by single-chain proteins instead of protein complexes, so there is no overlap between our training and test sets. We doubled check this by BLAST (34), which shows that none of the test protein pairs can be simultaneously aligned to a training protein.
RESULT
Evaluation and metrics
We compare ComplexContact with pure DCA methods such as CCMpred (11), Gremlin (35) and EVfold (36) and two related web servers GremlinComplex and EVcomplex. CCMpred, EVfold and Gremlin are pure co-evolution methods and their accuracy is not very different.
We evaluate the accuracy of the top L/5, 50, 20, 10, 5 predicted inter-protein contacts. The top-k accuracy is defined as the percentage of correct predictions among the top k predicted contacts. When the number of native contacts is smaller than k, we still use k as denominator in calculating accuracy, which may make the accuracy look small when k is big. More experimental results are available at the online documentation page http://raptorx2.uchicago.edu/ComplexContact/documentation/#6.
Performance on Baker and 3Dcomplex datasets
As shown in Table 1, tested on the Baker's dataset (of 32 protein pairs), ComplexContact greatly outperforms EVComplex (EVfold), GremlinComplex and CCMpred regardless of how many predicted contacts are evaluated.
Table 1. Inter-protein contact prediction accuracy (%) on Baker's dataset.
| Predictor | L/5 | 50 | 20 | 10 | 5 |
|---|---|---|---|---|---|
| EVcomplex(s) | 9.63 | 14.41 | 21.55 | 26.55 | 31.03 |
| GremlinComplex(s) | 14.67 | 26.00 | 41.21 | 52.76 | 58.62 |
| EVfold | 16.10 | 27.59 | 42.07 | 54.83 | 62.76 |
| CCMpred | 17.64 | 29.86 | 46.03 | 55.52 | 61.38 |
| ComplexContact (s) | 38.47 | 50.41 | 60.52 | 65.86 | 68.28 |
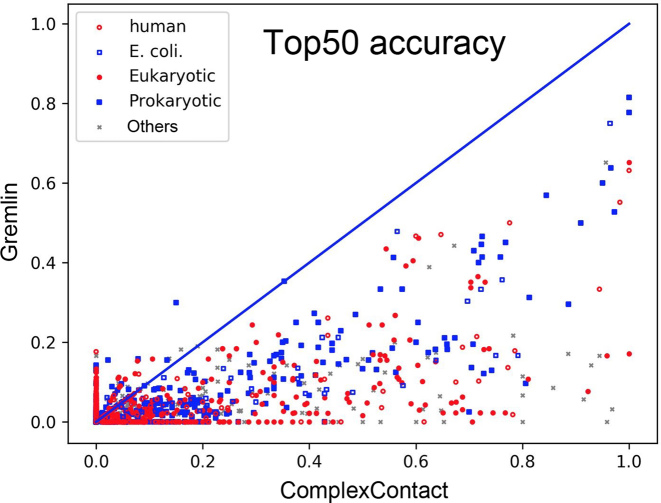
As shown in Figure 2, on a much larger benchmark (of 4479 heterodimers) extracted from 3D complex (37), ComplexContact outperforms Gremlin by a large margin regardless of the species of the test dimers.
Figure 2.
The top-50 prediction accuracy by ComplexContact and Gremlin on the protein pairs extracted from 3DComplex. Each dot represents one protein pair and is colored by its species. A dot below the diagonal line indicates that ComplexContact has a better accuracy.
The prediction accuracy depends on two main factors: the number of non-redundant sequence homologs in the multiple sequence alignment (MSA) and the interfacial contact density (measured by the number of interfacial contacts divided by sequence length sum). The former determines the quality of co-evolution signal (one of the input features of our DL method). The latter impacts prediction accuracy because our DL method makes use of contact occurrence patterns. When contact density is low, it is hard to identify reliable contact patterns.
Quality assessment of the predicted probability
ComplexContact predicts the probability of any two residues forming a contact. Here we assess the quality of the top 50 predicted probability values of the heterodimers extracted from 3Dcomplex.
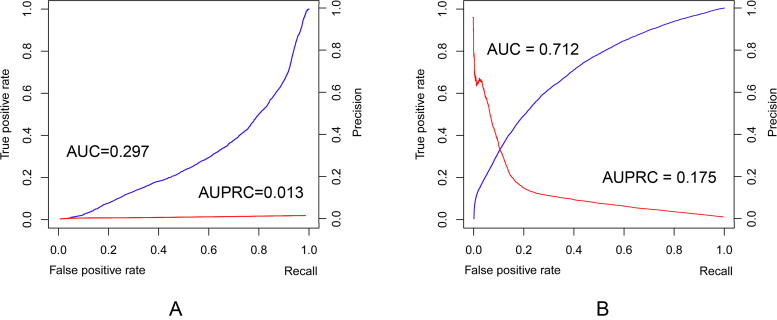
As shown in Figure 3, ComplexContact has much better AUC (Area Under the ROC curve) and AUPRC (Area Under the PR curve) (0.712 and 0.175, respectively, Figure 3B) than Gremlin (0.297 and 0.013, respectively, Figure 3A). Gremlin has an AUC <0.5, which implies that its contact selection is even worse than random guess. Table 2 shows the precision and recall for a list of probability values produced by ComplexContact. For example, when the predicted probability is >0.90, the precision is 0.57.
Figure 3.
Quality assessment of the top 50 predicted interfacial contacts for 4479 heterodimers extracted from 3Dcomplex. (A) and (B) show the precision-recall (in red) and ROC (in blue) curves generated by Gremlin and ComplexContact, respectively. AUC: Area Under the ROC curve; AUPRC: Area Under the precision-recall curve.
Table 2. Precision and recall of the top 50 predicted interfacial contacts by ComplexContact on the 3Dcomplex data.
| Probability | Precision | Recall |
|---|---|---|
| 0.95 | 0.70 | 0.03 |
| 0.90 | 0.57 | 0.06 |
| 0.85 | 0.45 | 0.09 |
| 0.80 | 0.35 | 0.12 |
| 0.75 | 0.27 | 0.14 |
| 0.70 | 0.22 | 0.17 |
| 0.65 | 0.19 | 0.21 |
| 0.60 | 0.16 | 0.27 |
| 0.55 | 0.14 | 0.35 |
| 0.50 | 0.13 | 0.44 |
SERVER IMPLEMENTATION
Overall description
Input
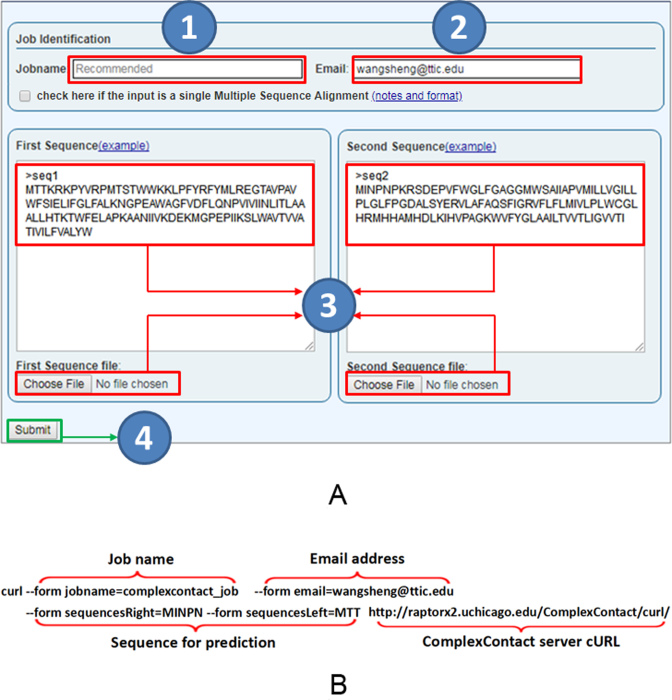
As shown in Figure 4, users may submit a single sequence pair, a pair of multiple sequence alignments or a batch of 20 sequence pairs by copying and pasting to the input text field or uploading files. A jobname and an email address are optional, but they can facilitate job retrieval.
Figure 4.
ComplexContact server job submission. (A) Users may submit a job by a web interface, which has fields for job name (1), optional user email address (2), and a pair of sequences (or multiple sequence alignments) (3). The sequences shall be in FASTA format and can also be submitted in a file. (B) Users may also submit a job by a publicly available program Curl without using the web interface. In this command, Job name and Email address are optional. A job URL will be returned on screen after submission. Curl allows users to submit a large number of jobs quickly.
Job retrieval
ComplexContact assigns one unique job ID and one URL to each submission for job retrieval. When an email is provided in submission, users will be notified by email once a batch of jobs are done; users may also retrieve their jobs by the ‘My Jobs’ link at the top right of the web page and by the ‘Job Status’ link, through which users may find a job by one of its submitted sequences.
Output
In addition to the original input sequences, the result web page has three result sections (see Figure 5). The first section visualizes the predicted contact map, which can be zoomed in and dragged around to facilitate detailed examination. Hovering mouse over the contact image will display the predicted contact probability value at a specific residue pair. The second section includes three panels: a panel for contact image zooming and dragging, a panel for downloading the predicted complex contact map, and a panel for downloading the detailed prediction results. The third section displays the two paired MSAs generated by genome- and phylogeny-based methods as well as the number of sequence homologs in each MSA.
Figure 5.
ComplexContact server result page. The left part shows the predicted complex contact map (1), where the predicted probability is displayed in greyscale, with a darker color indicating a larger value. The middle part shows three panels. The first one is used to zoom and drag contact images (2). The second panel is for downloading the predicted contact map (3), and the third panel is for downloading the detailed prediction results (4). The right part shows two paired MSAs generated by genome-based method (5) and phylogeny-based method (6).
Detailed prediction results
The downloadable file contains the followings: (a) the two input protein sequences in FASTA format; (b) MSAs generated by HHblits (38) for each input sequence; (c) two paired MSAs; (d) one predicted complex contact map for each paired MSA; (e) the final predicted contact map.
Processing time
The running time depends on the length of the two input sequences and the number of sequence homologs detected by the server. For a protein pair of ∼250 residues, it takes about one hour to finish one job after it is scheduled to run. When there are many waiting jobs (or jobs of long sequences) in the queue, it may take a few hours for a job to be scheduled to run. Nevertheless, most jobs can be done within one day after submission.
Documentation
The documentation of ComplexContact is available by the ‘Docs’ link at the web page. It includes some details about the server, descriptions of input and output, explanations of prediction results, a sample prediction result and more experimental results.
CONCLUSION AND FUTURE WORK
We have presented ComplexContact, a web server for sequence-based interfacial residue-residue contact prediction using deep learning and residue co-variation. ComplexContact outperforms similar servers by a large margin regardless of the species. However, it shall be noted that complex contact prediction is a very challenging problem and there is still a large room for improvement. Currently our DL model was trained using only single-chain proteins. According to our experience on membrane protein contact prediction (39), using a mix of membrane and soluble proteins to train a DL model works better than using soluble proteins or membrane proteins alone. Therefore, we plan to further improve interfacial contact prediction accuracy by training a DL model using a mix of single-chain proteins and protein complexes.
FUNDING
National Institutes of Health (NIH) [R01GM089753 to J.X.]; National Science Foundation (NSF) [DBI-1564955 to J.X.]. Funding for open access charge: NIH; NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jones S., Thornton J.M.. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lensink M.F., Velankar S., Wodak S.J.. Modeling protein–protein and protein–peptide complexes: CAPRI 6th edition. Proteins: Struct. Funct. Bioinform. 2017; 85:359–377. [DOI] [PubMed] [Google Scholar]
- 3.Mosca R., Céol A., Aloy P.. Interactome3D: adding structural details to protein networks. Nat. Methods. 2013; 10:47. [DOI] [PubMed] [Google Scholar]
- 4.Ovchinnikov S., Kamisetty H., Baker D.. Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. Elife. 2014; 3:e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopf T.A., Schärfe C.P., Rodrigues J.P., Green A.G., Kohlbacher O., Sander C., Bonvin A.M., Marks D.S.. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014; 3:e03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marks D.S., Hopf T.A., Sander C.. Protein structure prediction from sequence variation. Nat. Biotechnol. 2012; 30:1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigt M., White R.A., Szurmant H., Hoch J.A., Hwa T.. Identification of direct residue contacts in protein–protein interaction by message passing. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Juan D., Pazos F., Valencia A.. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013; 14:249. [DOI] [PubMed] [Google Scholar]
- 9.Ma J., Wang S., Wang Z., Xu J.. Protein contact prediction by integrating joint evolutionary coupling analysis and supervised learning. Bioinformatics. 2015; 31:3506–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones D.T., Buchan D.W., Cozzetto D., Pontil M.. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2011; 28:184–190. [DOI] [PubMed] [Google Scholar]
- 11.Seemayer S., Gruber M., Söding J.. CCMpred—fast and precise prediction of protein residue–residue contacts from correlated mutations. Bioinformatics. 2014; 30:3128–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Li W., Zhang R., Liu S., Xu J.. CoinFold: a web server for protein contact prediction and contact-assisted protein folding. Nucleic Acids Res. 2016; 44:W361–W366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gueudré T., Baldassi C., Zamparo M., Weigt M., Pagnani A.. Simultaneous identification of specifically interacting paralogs and interprotein contacts by direct coupling analysis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:12186–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaján L., Hopf T.A., Kalaš M., Marks D.S., Rost B.. FreeContact: fast and free software for protein contact prediction from residue co-evolution. BMC Bioinformatics. 2014; 15:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morcos F., Pagnani A., Lunt B., Bertolino A., Marks D.S., Sander C., Zecchina R., Onuchic J.N., Hwa T., Weigt M.. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Sun S., Li Z., Zhang R., Xu J.. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 2017; 13:e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adhikari B., Hou J., Cheng J.. DNCON2: Improved protein contact prediction using two-level deep convolutional neural networks. Bioinformatics. 2017; 34:1466–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhikari B., Hou J., Cheng J.. Protein contact prediction by integrating deep multiple sequence alignments, coevolution and machine learning. Proteins: Struct. Funct. Bioinform. 2018; 86:84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl K., Schneider M., Brock O.. EPSILON-CP: using deep learning to combine information from multiple sources for protein contact prediction. BMC Bioinformatics. 2017; 18:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong D., Zeng J., Gong H.. A deep learning framework for improving long-range residue–residue contact prediction using a hierarchical strategy. Bioinformatics. 2017; 33:2675–2683. [DOI] [PubMed] [Google Scholar]
- 21.Du T., Liao L., Wu C.H., Sun B.. Prediction of residue-residue contact matrix for protein-protein interaction with Fisher score features and deep learning. Methods. 2016; 110:97–105. [DOI] [PubMed] [Google Scholar]
- 22.He B., Mortuza S., Wang Y., Shen H.-B., Zhang Y.. NeBcon: protein contact map prediction using neural network training coupled with naïve Bayes classifiers. Bioinformatics. 2017; 33:2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skwark M.J., Raimondi D., Michel M., Elofsson A.. Improved contact predictions using the recognition of protein like contact patterns. PLoS Comput. Biol. 2014; 10:e1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S., Sun S., Xu J.. Analysis of deep learning methods for blind protein contact prediction in CASP12. Proteins: Struct. Funct. Bioinform. 2018; 86:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaarschmidt J., Monastyrskyy B., Kryshtafovych A., Bonvin A.M.. Assessment of contact predictions in CASP12: co-evolution and deep learning coming of age. Proteins: Struct. Funct. Bioinform. 2018; 86:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou T., Wang S., Xu J.. Deep learning reveals many more inter-protein residue-residue contacts than direct coupling analysis. Lect. Notes Comput. Sci. 2017; 295–296.LNBI 10812.
- 27.Feinauer C., Szurmant H., Weigt M., Pagnani A.. Inter-protein sequence co-evolution predicts known physical interactions in bacterial ribosomes and the Trp operon. PLoS One. 2016; 11:e0149166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H., Zhou M., Alkins R.. Group role assignment via a Kuhn–Munkres algorithm-based solution. IEEE Trans. Syst. Man Cybernet.-Part A: Syst. Hum. 2012; 42:739–750. [Google Scholar]
- 29.Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2011; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He K., Zhang X., Ren S., Sun J.. Deep residual learning for image recognition. Proc. IEEE Conf. Comput. Vis. Pattern Recogni. 2016; 770–778. [Google Scholar]
- 31.Wang S., Peng J., Ma J., Xu J.. Protein secondary structure prediction using deep convolutional neural fields. Scientific Rep. 2016; 6:18962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J., Wang S.. AcconPred: Predicting solvent accessibility and contact number simultaneously by a multitask learning framework under the conditional neural fields model. BioMed Res. Int. 2015; 2015:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S., Li W., Liu S., Xu J.. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 2016; 44:W430–W435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamisetty H., Ovchinnikov S., Baker D.. Assessing the utility of coevolution-based residue–residue contact predictions in a sequence-and structure-rich era. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks D.S., Colwell L.J., Sheridan R., Hopf T.A., Pagnani A., Zecchina R., Sander C.. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011; 6:e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy E.D., Pereira-Leal J.B., Chothia C., Teichmann S.A.. 3D complex: a structural classification of protein complexes. PLoS Comput. Biol. 2006; 2:e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remmert M., Biegert A., Hauser A., Söding J.. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012; 9:173. [DOI] [PubMed] [Google Scholar]
- 39.Wang S., Li Z., Yu Y., Xu J.. Folding membrane proteins by deep transfer learning. Cell Syst. 2017; 5:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iserte J., Simonetti F.L., Zea D.J., Teppa E., Marino-Buslje C.. I-COMS: Interprotein-COrrelated Mutations Server. Nucleic Acids Res. 2015; 43:W320–W325. [DOI] [PMC free article] [PubMed] [Google Scholar]