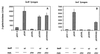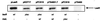RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa - PubMed (original) (raw)
RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa
T de Kievit et al. J Bacteriol. 1999 Apr.
Abstract
As components of a Pseudomonas aeruginosa quorum-sensing system, LasR and PAI-1 globally regulate expression of multiple virulence determinants, as well as the second P. aeruginosa quorum-sensing system. To date, no information exists on negative regulation of the quorum-sensing cascade in P. aeruginosa. Here we describe a novel gene, rsaL, which is located downstream from lasR and transcribed antisense relative to lasR. In P. aeruginosa, overexpression of rsaL results in reduced lasB expression and decreased elastase activity. With the use of a six-His protein fusion system, we demonstrate that rsaL encodes an 11-kDa protein. Direct quantitation of PAI-1 levels in cultures and studies utilizing Escherichia coli lambda lysogens carrying lacZ transcriptional fusions reveal that RsaL specifically represses transcription of the PAI-1 autoinducer synthase gene, lasI. RsaL's repressive effect on lasI and the associated decrease in elastase activity have important implications for the expression of all LasR-PAI-1-dependent virulence genes and the overall pathogenicity of P. aeruginosa.
Figures
FIG. 1
Plasmid inserts and DNA sequence of the rsaL gene. (A) The region including and surrounding the lasR, rsaL, and lasI genes is depicted. Restriction sites include _Eco_RV (RV), _Eco_RI (E; E* indicates that this restriction site was located in the multiple cloning site of vector pLP170 [29] and not in P. aeruginosa DNA), _Sfi_I (S), and _Hph_I (H). The 658-bp _Eco_RI DNA insert found on pPCS2001 and pPCS2002 is indicated. The 649-bp Eco_RV fragment that was deleted and replaced with a tetracycline resistance (Tetr) cassette during the generation of strain PAO-R1 (Δ_lasR) (9) is also shown. The 307-bp _Sfi_I-_Hph_I fragment encodes a functional RsaL protein; thus, the Tetr cassette insertion in strain PAO-R1 does not interrupt the rsaL gene. (B) DNA inserts of the plasmids used in this study are indicated. a, plasmids containing both lasR and rsaL in their native conformation on a single DNA fragment; b, plasmids that contain the lasR and rsaL genes on separate DNA fragments. (C) lasR-lasI intergenic region. The deduced amino acid sequence of RsaL is indicated in bold. The rsaL translational start codon (ATG) is underlined, and potential _lux_-box-like elements are enclosed by boxes.
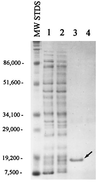
FIG. 2
Expression of RsaL as a fusion protein. Lysates from E. coli JM109 carrying either the control vector (pTrcHisB) (lane 1) or the vector with an in-frame fusion of rsaL (pTrcHisB/rsaL) (lane 2) were separated by SDS-PAGE. Ni-NTA resin was used to purify the fusion protein from E. coli JM109(pTrcHisB/rsaL) (lane 3) and from E. coli JM109 carrying rsaL cloned onto pTrcHisA (lane 4) to serve as an out-of-frame control. Molecular mass standards are shown in daltons. The arrow indicates the protein observed in lane 3, which is believed to be RsaL.
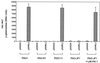
FIG. 3
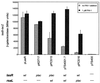
Effect of the P. aeruginosa quorum-sensing regulators on expression of rsaL. Strains carrying a plasmid-borne rsaL-lacZ translational fusion (pSWRL2) or the control plasmid (pSW205) were monitored for expression of β-galactosidase. Expression of the fusion was analyzed in the wild-type strain PAO1, the LasR null mutant PAO-R1, the RhlR null mutant PDO111, and the LasI null mutant PAO-JP1. PAO-JP1 grown in the presence of 1 μM exogenous PAI-1 was also examined.
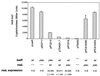
FIG. 4
Overexpression of rsaL inhibits expression of lasB in P. aeruginosa. β-Galactosidase activity from a plasmid-borne lasB-lacZ translational fusion was monitored in the presence of either lasR alone or both lasR and rsaL in P. aeruginosa PAO-R1. Expression of lasR and rsaL was directed by either their native promoters (wt) or the lac promoter (plac) as indicated under each plasmid. Plasmids in which the RsaL start methionine is removed (pPCS16/NS) or in which a stop codon is prematurely introduced (pPCS16/ES), but which otherwise are identical to pPCS16, were also examined for β-galactosidase production. To monitor the level of rsaL expression in plasmids pPCS15, pTS4001.7, and pPCS16, the rsaL gene located on these constructs was replaced by an rsaL-lacZ translational fusion. β-Galactosidase activity from the rsaL-lacZ fusions in P. aeruginosa PAO-R1 is indicated under each plasmid. n.d., not determined.
FIG. 5
Effect of RsaL on lasI expression. The expression of lasB-lacZ (A) and lasI-lacZ (B) fusions carried as prophages in E. coli MG4 was examined in the presence of lasR under control of its own promoter (pEXR), rsaL overexpressed from the tac promoter (pEXRL), or lasR expressed from its own promoter and rsaL overexpressed from the tac promoter (pEXRR). A construct identical to pEXRR but which has the start codon of RsaL removed through mutagenesis (pEXRR/NS) was also examined. Plasmid pEX1.8 was used as a control vector in these experiments. In all cases, 100 nM PAI-1 and 1 mM IPTG were added to the cultures at the initiation of growth.
FIG. 6
RsaL’s repressive effect is overcome by addition of exogenous PAI-1. β-Galactosidase activity from a plasmid-borne lasB-lacZ translational fusion was monitored in the presence of either lasR alone or both lasR and rsaL in P. aeruginosa PAO-R1 after growth in either the presence or absence of 1 μM exogenous PAI-1. Expression of lasR and rsaL was directed by either their native promoters (wt) or the lac promoter (plac) as indicated under each plasmid.
FIG. 7
Western blot analysis of LasR expression when rsaL is overexpressed in P. aeruginosa PAO-R1. Anti-LasR antibody (1:5,000) was used to visualize LasR in total cell lysates of strain PAO-R1 in the presence of lasR alone or both lasR and rsaL. Expression of lasR and rsaL was directed by either their native promoters (wt) or the lac promoter (plac) as indicated.
Similar articles
- Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy.
Seed PC, Passador L, Iglewski BH. Seed PC, et al. J Bacteriol. 1995 Feb;177(3):654-9. doi: 10.1128/jb.177.3.654-659.1995. J Bacteriol. 1995. PMID: 7836299 Free PMC article. - Analysis of the organization and expression patterns of the convergent Pseudomonas aeruginosa lasR/rsaL gene pair uncovers mutual influence.
Schinner S, Preusse M, Kesthely C, Häussler S. Schinner S, et al. Mol Microbiol. 2021 Apr;115(4):643-657. doi: 10.1111/mmi.14628. Epub 2020 Nov 11. Mol Microbiol. 2021. PMID: 33073409 - A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa.
Pearson JP, Passador L, Iglewski BH, Greenberg EP. Pearson JP, et al. Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1490-4. doi: 10.1073/pnas.92.5.1490. Proc Natl Acad Sci U S A. 1995. PMID: 7878006 Free PMC article. - An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR.
Chugani S, Greenberg EP. Chugani S, et al. Front Cell Infect Microbiol. 2014 Oct 28;4:152. doi: 10.3389/fcimb.2014.00152. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25389523 Free PMC article. Review. - Growing repertoire of AraC/XylS activators.
Egan SM. Egan SM. J Bacteriol. 2002 Oct;184(20):5529-32. doi: 10.1128/JB.184.20.5529-5532.2002. J Bacteriol. 2002. PMID: 12270809 Free PMC article. Review. No abstract available.
Cited by
- Bacterial quorum sensing: its role in virulence and possibilities for its control.
Rutherford ST, Bassler BL. Rutherford ST, et al. Cold Spring Harb Perspect Med. 2012 Nov 1;2(11):a012427. doi: 10.1101/cshperspect.a012427. Cold Spring Harb Perspect Med. 2012. PMID: 23125205 Free PMC article. Review. - The atypical organization of the luxI/R family genes in AHL-driven quorum-sensing circuits.
Cai Y, Zhang X. Cai Y, et al. J Bacteriol. 2024 Feb 22;206(2):e0043023. doi: 10.1128/jb.00430-23. Epub 2024 Jan 19. J Bacteriol. 2024. PMID: 38240569 Free PMC article. Review. - Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1.
Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. Heurlier K, et al. J Bacteriol. 2003 Apr;185(7):2227-35. doi: 10.1128/JB.185.7.2227-2235.2003. J Bacteriol. 2003. PMID: 12644493 Free PMC article. - Look who's talking: communication and quorum sensing in the bacterial world.
Williams P, Winzer K, Chan WC, Cámara M. Williams P, et al. Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1119-34. doi: 10.1098/rstb.2007.2039. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360280 Free PMC article. Review. - The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II.
Dubern JF, Lugtenberg BJ, Bloemberg GV. Dubern JF, et al. J Bacteriol. 2006 Apr;188(8):2898-906. doi: 10.1128/JB.188.8.2898-2906.2006. J Bacteriol. 2006. PMID: 16585751 Free PMC article.
References
- Bassler B L, Wright M, Silverman M R. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12:403–412. - PubMed
- Cui Y, Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol. 1995;177:5108–5115. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous