Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein - PubMed (original) (raw)
Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein
W Yuan et al. Mol Biol Cell. 1999 May.
Free PMC article
Abstract
Cytosolic and peroxisomal enzymes necessary for methanol assimilation are synthesized when Pichia pastoris is grown in methanol. Upon adaptation from methanol to a glucose environment, these enzymes are rapidly and selectively sequestered and degraded within the yeast vacuole. Sequestration begins when the vacuole changes shape and surrounds the peroxisomes. The opposing membranes then fuse, engulfing the peroxisome. In this study, we have characterized a mutant cell line (glucose-induced selective autophagy), gsa7, which is defective in glucose-induced selective autophagy of peroxisomes, and have identified the GSA7 gene. Upon glucose adaptation, gsa7 cells were unable to degrade peroxisomal alcohol oxidase. We observed that the peroxisomes were surrounded by the vacuole, but complete uptake into the vacuole did not occur. Therefore, we propose that GSA7 is not required for initiation of autophagy but is required for bringing the opposing vacuolar membranes together for homotypic fusion, thereby completing peroxisome sequestration. By sequencing the genomic DNA fragment that complemented the gsa7 phenotype, we have found that GSA7 encodes a protein of 71 kDa (Gsa7p) with limited sequence homology to a family of ubiquitin-activating enzymes, E1. The knockout mutant gsa7Delta had an identical phenotype to gsa7, and both mutants were rescued by an epitope-tagged Gsa7p (Gsa7-hemagglutinin [HA]). In addition, a GSA7 homolog, APG7, a protein required for autophagy in Saccharomyces cerevisiae, was capable of rescuing gsa7. We have sequenced the human homolog of GSA7 and have shown many regions of identity between the yeast and human proteins. Two of these regions align to the putative ATP-binding domain and catalytic site of the family of ubiquitin activating enzymes, E1 (UBA1, UBA2, and UBA3). When either of these sites was mutated, the resulting mutants [Gsa7(DeltaATP)-HA and Gsa7(C518S)-HA] were unable to rescue gsa7 cells. We provide evidence to suggest that Gsa7-HA formed a thio-ester linkage with a 25-30 kDa protein. This conjugate was not observed in cells expressing Gsa7(DeltaATP)-HA or in cells expressing Gsa7(C518S)-HA. Our results suggest that this unique E1-like enzyme is required for homotypic membrane fusion, a late event in the sequestration of peroxisomes by the vacuole.
Figures
Figure 1
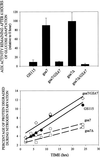
Peroxisome degradation during nutrient adaptation and protein degradation during nitrogen starvation in parental GS115 and gsa7–1 mutant. Cells were grown for 24–36 h in methanol induction medium. At 0 and 6 h of glucose (2%) adaptation, cell-free extracts were prepared, and alcohol oxidase (AOX) and formate dehydrogenase (FDH) activities were assayed as described in MATERIALS AND METHODS. The values represent the mean ± SD of three or more determinations and are presented as a percentage of the activity measured at 0 h. The degradation of cellular proteins during nitrogen starvation was measured as described in MATERIALS AND METHODS. Cellular proteins were radiolabeled and then chased in medium containing histidine and nitrogen (Fed) or lacking amino acids and nitrogen (Starved). The production of TCA-soluble radioactivity was measured during 2–24 h of chase, and the rates of degradation were determined by linear regression. The values are presented as the percentage degraded per hour ± SE of the linear regression.
Figure 2
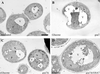
Morphology of gsa7 and _gsa7_Δ cells during glucose adaptation. _gsa7_Δ cells were grown in methanol (A) and gsa7, _gsa7_Δ, and _gsa7_Δ cells transformed with the normal GSA7 gene (gsa7Δ/GSA7) were grown in methanol induction medium then adapted to glucose for 3 h (B–D). Cells were harvested, fixed in potassium permanganate, and prepared for electron microscopy as described in MATERIALS AND METHODS. N, nucleus; V, vacuole; P, peroxisome. Bar, 0.5 μm.
Figure 3
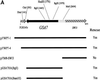
Nucleotide and predicted amino acid sequence of GSA7. gsa7 cells were stably transformed with pYWP7-1 (containing a 4.5-kb genomic DNA insert), pYWP7-4 (containing a 4.1-kb genomic DNA insert), pGSA7HA (containing a 2.3-kb ORF of GSA7 with an HA epitope at its C terminus), and pWP-SWI3 (containing a 1.5-kb 3′ fragment of the original 4.1-kb genomic DNA). pGSA7HA(_Bgl_I) was linearized within the _GSA7_gene with _Bgl_I to promote insertion into the gsa7 locus. pGSA7HA (_Bam_HI) was linearized within the HIS4 gene with _Bam_HI to promote insertion into the his4 locus. The resulting transformants were analyzed for their ability to rescue the mutant phenotype as described previously (Yuan et al., 1997). The GSA7 ORF encodes a protein of 989 amino acids with a molecular mass of 71 kDa. The GSA7 sequence has a GenBank accession number of AF098976.
Figure 3
Nucleotide and predicted amino acid sequence of GSA7. gsa7 cells were stably transformed with pYWP7-1 (containing a 4.5-kb genomic DNA insert), pYWP7-4 (containing a 4.1-kb genomic DNA insert), pGSA7HA (containing a 2.3-kb ORF of GSA7 with an HA epitope at its C terminus), and pWP-SWI3 (containing a 1.5-kb 3′ fragment of the original 4.1-kb genomic DNA). pGSA7HA(_Bgl_I) was linearized within the _GSA7_gene with _Bgl_I to promote insertion into the gsa7 locus. pGSA7HA (_Bam_HI) was linearized within the HIS4 gene with _Bam_HI to promote insertion into the his4 locus. The resulting transformants were analyzed for their ability to rescue the mutant phenotype as described previously (Yuan et al., 1997). The GSA7 ORF encodes a protein of 989 amino acids with a molecular mass of 71 kDa. The GSA7 sequence has a GenBank accession number of AF098976.
Figure 4
Rescue of gsa7 and _gsa7_Δ by Gsa7p. gsa7 and _gsa7_Δ cells were stably transformed with pGSA7HA(_Bam_HI). Those colonies that grew on His− medium were screened for expression of Gsa7-HA by Western blotting with an antibody to the HA epitope. The nontransformed (gsa7 and _gsa7_Δ) and transformed (gsa7/GSA7 and _gsa7_Δ/GSA7) cells were then grown in methanol induction medium and adapted to glucose for 6 h. The AOX activities are presented as a percentage of the activities measured at 0 h and represent the mean ± SD of four or more determinations. The degradation of cellular proteins during nitrogen starvation was performed as described previously in MATERIALS AND METHODS. Cellular proteins were radiolabeled and then chased in starvation medium lacking histidine and nitrogen. The production of TCA-soluble radioactivity was measured at 2–24 h of chase.
Figure 5
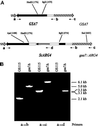
Preparation and characterization of the GSA7 knockout. The S. cerevisiae ARG4 gene was inserted into _Hin_dIII and _Bgl_II sites within the ORF of GSA7. The ARG4 insert included the ORF of ARG4 (arrow) and 1300 bp of 5′ and 500 bp of 3′ noncoding regions. The 5′ noncoding region includes the ARG4 promoter, whereas the 3′ noncoding region presumably lacks a promoter sequence, because the next gene downstream of ARG4 is 2200 bp away. In addition, the 3′ noncoding region likely contains a transcription terminator. The knockout fragment (5.1 kb) was cut out of the shuttle vector by _Apa_I and _Sca_I digestion and used to transform PPF1 (his4, arg4). Stable Arg+ transformants were isolated, gsa mutants were identified by direct colony assay, and the recombination of this construct with the genomic DNA was confirmed by PCR. Genomic DNA was isolated from GS115 and _gsa7_Δ, and the site of insertion of the ARG4 gene was verified by PCR analysis using the primers inside (b and d) and outside (a and c) the insert. The expected fragments of 5.0 kb (a → b), 6.1 kb (a → c), and 3.3 kb (a → d) were consistent with the ARG4 gene being inserted into the GSA7 locus of _gsa7_Δ cells.
Figure 6
Comparison of GSA7 genes. The amino acid sequence of GSA_7 from P. pastoris (see Figure 3B) was aligned to APG7 from S. cerevisiae, and GSA7 was aligned from H. sapiens (Hs_GSA7). The sequence of Hs_GSA7_ has a GenBank accession number of AF094516. Amino acid identity represented by dashed lines was evident within many regions of the sequence. Gaps represented by dots were inserted to optimize amino acid alignments.
Figure 7
Complementation of gsa7 by APG7. gsa7 cells were stably transformed with pGSA7HA(_Bam_HI) (lanes A and B), pGSA7HA(_Bgl_I) (lane C), or pAPG7HA(_Bam_HI) (lanes D–F) behind their respective endogenous promoters. Expression of Gsa7-HA and Apg7-HA was evaluated by Western blotting with a monoclonal antibody to the HA epitope. Clones expressing and nonexpressing Gsa7-HA and Apg7-HA were grown in methanol induction medium and adapted to glucose for 6 h. Cells were lysed, and the extracts were assayed for AOX activity (see MATERIALS AND METHODS). The numbers represent the percentage of AOX activity present at 6 h relative to that measured at 0 h.
Figure 8
Complementation of gsa7 by normal and mutant forms of recombinant Gsa7-HA. (A) GSA7, APG7, and HsGSA7 proteins were aligned to three ubiquitin-activating enzymes (UBA1, UBA2, and UBA3). The amino acid alignment around the putative catalytic domain (C518) and ATP-binding region (K327–R342) are indicated. Amino acid identity represented by dashed lines was evident within these regions. Gaps represented by dots were inserted to optimize amino acid alignments. (B) gsa7 cells were stably transformed with GSA7-HA, _gsa7(Δ_ATP), gsa7(C518S), and gsa7(C562S). The cells were then grown in methanol induction medium then adapted to glucose for 6 h. Cell extracts were prepared, and AOX assays were performed. The resulting values are presented as a percentage of the activity measured at 0 h and represent the mean ± SD of four or more determinations. (C) Aliquots of cell extracts prepared at 3 h of glucose adaptation were solubilized in 2% SDS and boiled for 3 min (−DTT) or solubilized in 2% SDS with 1.5% DTT and boiled for 5 min (+DTT), and the proteins were separated by SDS-PAGE. After transfer to nitrocellulose, the epitope-tagged Gsa7 proteins were identified using polyclonal antibodies that recognized the HA epitope. Gsa7-HA migrated as a 72-kDa protein (arrow). A thio-ester conjugate of Gsa7-HA was observed at ∼100 kDa (arrowhead).
Similar articles
- Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase.
Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA Jr. Yuan W, et al. J Cell Sci. 1997 Aug;110 ( Pt 16):1935-45. doi: 10.1242/jcs.110.16.1935. J Cell Sci. 1997. PMID: 9296392 - Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris.
Tuttle DL, Dunn WA Jr. Tuttle DL, et al. J Cell Sci. 1995 Jan;108 ( Pt 1):25-35. doi: 10.1242/jcs.108.1.25. J Cell Sci. 1995. PMID: 7738102 - GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris.
Strømhaug PE, Bevan A, Dunn WA Jr. Strømhaug PE, et al. J Biol Chem. 2001 Nov 9;276(45):42422-35. doi: 10.1074/jbc.M104087200. Epub 2001 Aug 31. J Biol Chem. 2001. PMID: 11533052 - Pexophagy: the selective autophagy of peroxisomes.
Dunn WA Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Dunn WA Jr, et al. Autophagy. 2005 Jul;1(2):75-83. doi: 10.4161/auto.1.2.1737. Epub 2005 Jul 13. Autophagy. 2005. PMID: 16874024 Review. - Peroxisome biogenesis in Saccharomyces cerevisiae.
Kunau WH, Hartig A. Kunau WH, et al. Antonie Van Leeuwenhoek. 1992 Aug;62(1-2):63-78. doi: 10.1007/BF00584463. Antonie Van Leeuwenhoek. 1992. PMID: 1444337 Review.
Cited by
- The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation.
Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. Suzuki K, et al. EMBO J. 2001 Nov 1;20(21):5971-81. doi: 10.1093/emboj/20.21.5971. EMBO J. 2001. PMID: 11689437 Free PMC article. - Autophagy as a regulated pathway of cellular degradation.
Klionsky DJ, Emr SD. Klionsky DJ, et al. Science. 2000 Dec 1;290(5497):1717-21. doi: 10.1126/science.290.5497.1717. Science. 2000. PMID: 11099404 Free PMC article. Review. - Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae.
Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Roberts P, et al. Mol Biol Cell. 2003 Jan;14(1):129-41. doi: 10.1091/mbc.e02-08-0483. Mol Biol Cell. 2003. PMID: 12529432 Free PMC article. - Nitrogen Starvation and Stationary Phase Lipophagy Have Distinct Molecular Mechanisms.
Kumar R, Rahman MA, Nazarko TY. Kumar R, et al. Int J Mol Sci. 2020 Nov 29;21(23):9094. doi: 10.3390/ijms21239094. Int J Mol Sci. 2020. PMID: 33260464 Free PMC article. - Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways.
George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. George MD, et al. Mol Biol Cell. 2000 Mar;11(3):969-82. doi: 10.1091/mbc.11.3.969. Mol Biol Cell. 2000. PMID: 10712513 Free PMC article.
References
- Ahlberg L, Berkenstam A, Henell F, Glaumann H. Degradation of short and long lived proteins in isolated rat liver lysosomes: effects of pH, temperature, and proteolytic inhibitors. J Biol Chem. 1985;260:5847–5854. - PubMed
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1988.
- Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the antiestrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous