La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA - PubMed (original) (raw)
La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA
T Heise et al. J Virol. 1999 Jul.
Abstract
We recently identified three nuclear proteins (p45, p39, and p26) that bind to a 91-nucleotide (nt) RNA element between nt 1243 and 1333 in hepatitis B virus (HBV) RNA, and we showed that these proteins and HBV RNA are regulated coordinately by gamma interferon and tumor necrosis factor alpha. Purification and sequence analysis of tryptic peptides obtained from p39 revealed sequence homology to the mouse La protein. Immunoprecipitation experiments showed that p45, p39, and p26 were recognized by anti-La-specific antiserum, indicating that p45 is the full-length La protein and that p39 and p26 are likely to be proteolytic La cleavage products. Furthermore, in competition experiments we found that all three La proteins bind, in a phosphorylation-dependent manner, to the same predicted stem-loop structure located between nt 1275 and 1291 of HBV, with Kds of approximately 1.0 nM. Collectively, these results support the notion that the La protein may contribute to HBV RNA stability, constitutively and in response to inflammatory cytokines.
Figures
FIG. 1
HBV RNA-binding proteins p45 and p39 are detectable in liver nuclear extracts from NaCl-injected mice, while p39 and p26 are detectable in CTL-injected mice. (A) Northern blotting and UV cross-linking analysis of 20 μg of total liver RNA or 5 μg of liver nuclear extract prepared from the same liver were performed as described in Materials and Methods. Sex and serum HBsAg-matched mice (lineage 1.3.32) were intravenously injected with 107 CTLs or with saline and sacrificed on day 5 after CTL administration. The upper panel shows the Northern blot analysis, and the lower panel shows the UV cross-linking analysis of nuclear extracts. (B) Predicted secondary structure of HBV in vitro transcript RNA.B used in this study. The secondary structure was calculated with the program MFOLD version 3 by Zuker and Turner available on the MFOLD server (71, 74). Arrows indicate the 3′ ends of in vitro transcripts RNA.C and RNA.D and of an oligoribonucleotide, RNA.E. The positions for all RNAs are shown according to the HBV ayw subtype sequence.
FIG. 2
Sequence analysis of tryptic peptides obtained from p39 revealed 100% homology to the mouse La protein. p39 was purified and processed as described in Materials and Methods. The sequence tags observed by N-terminal sequencing of three tryptic peptides obtained from p39 are shown in boldface.
FIG. 3
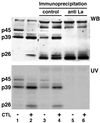
HBV RNA-binding proteins are recognized by anti-La-positive human serum. Five micrograms of liver nuclear extract prepared from untreated or CTL-injected mice (lanes 1 and 2) and 5 μg of liver nuclear extract from untreated or CTL-injected mice after immunoprecipitation with control human serum (lanes 3 and 4) or with anti-La-positive human serum (lanes 5 and 6) were incubated with 40 fmol of in vitro-labeled RNA.B. The UV cross-linking reaction was performed as described in Materials and Methods. The gel was transferred to a nitrocellulose membrane and analyzed for La protein by Western blotting (WB) and by autoradiography to detect p45, p39, and p26 as described in Materials and Methods.
FIG. 4
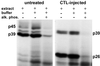
RNA-binding activity of p45, p39, and p26 depends on their phosphorylation status. Two micrograms of liver nuclear extract from untreated or CTL-injected mice was treated with 0.5 and 1.0 U of CIAP (alk. phos.) prior to addition of 40 fmol of 32P-labeled in vitro transcript RNA.B. The dephosphorylation reaction was performed in 20 μl of 1× reaction buffer for 30 min at 37°C. The binding reaction was performed and analyzed as described in Materials and Methods.
FIG. 5
Competition of various in vitro transcripts with RNA.B for binding of p45, p39, and p26. UV cross-linking experiments were performed with liver nuclear extracts from untreated or CTL-injected mice under standard conditions described in Materials and Methods. A 30- and a 60-fold molar excess of unlabeled in vitro transcripts RNA.B, GAPDH, actin, RRE, and CTE were added into the binding reaction mixture prior to the addition of 40 fmol of 32P-labeled RNA.B.
FIG. 6
Mapping of RNA.B for a sequential-structural element recognized by p45, p39, and p26. UV cross-linking experiments were performed with liver nuclear extracts from untreated or CTL-injected mice under standard conditions described in Materials and Methods. A 30- and a 60-fold molar excess of unlabeled in vitro transcripts RNA.B, RNA.C, RNA.D, and RNA.E (Fig. 1B) were added into the binding reaction mixture prior to the addition of 40 fmol of 32P-labeled RNA.B.
FIG. 7
Sequential and/or structural features of stem-loop 2 are substantive for the binding of p45, p39, and p26. UV cross-linking experiments were performed with liver nuclear extracts from untreated or CTL-injected mice under standard conditions described in Materials and Methods. A 10- and a 30-fold molar excess of unlabeled in vitro transcripts RNA.D-M1 to RNA.D-M6 were added into the binding reaction mixture prior to the addition of 40 fmol of 32P-labeled RNA.B. The decreases in signal intensity for p45, p39, and p26 were separately analyzed by phosphorimaging. The upper panel shows the predicted structure of RNA.D with nucleotide changes indicated by the arrows. The lower panel shows the plot of complex formation (arbitrary units) versus competitor concentrations (10- and 30-fold). WT, wild type.
Similar articles
- Characterization of nuclear RNases that cleave hepatitis B virus RNA near the La protein binding site.
Heise T, Guidotti LG, Chisari FV. Heise T, et al. J Virol. 2001 Aug;75(15):6874-83. doi: 10.1128/JVI.75.15.6874-6883.2001. J Virol. 2001. PMID: 11435567 Free PMC article. - Molecular characterization of the human La protein.hepatitis B virus RNA.B interaction in vitro.
Horke S, Reumann K, Rang A, Heise T. Horke S, et al. J Biol Chem. 2002 Sep 20;277(38):34949-58. doi: 10.1074/jbc.M201911200. Epub 2002 Jul 16. J Biol Chem. 2002. PMID: 12121976 - Functional characterization of the interaction between human La and hepatitis B virus RNA.
Ehlers I, Horke S, Reumann K, Rang A, Grosse F, Will H, Heise T. Ehlers I, et al. J Biol Chem. 2004 Oct 15;279(42):43437-47. doi: 10.1074/jbc.M402227200. Epub 2004 Aug 9. J Biol Chem. 2004. PMID: 15302879 - Hepatitis B virus nuclear export elements: RNA stem-loop α and β, key parts of the HBV post-transcriptional regulatory element.
Lim CS, Brown CM. Lim CS, et al. RNA Biol. 2016 Sep;13(9):743-7. doi: 10.1080/15476286.2016.1166330. Epub 2016 Mar 31. RNA Biol. 2016. PMID: 27031749 Free PMC article. Review. - La protein and its associated small nuclear and nucleolar precursor RNAs.
Maraia RJ, Intine RV. Maraia RJ, et al. Gene Expr. 2002;10(1-2):41-57. Gene Expr. 2002. PMID: 11868987 Free PMC article. Review.
Cited by
- HBV pathogenesis in animal models: recent advances on the role of platelets.
Iannacone M, Sitia G, Ruggeri ZM, Guidotti LG. Iannacone M, et al. J Hepatol. 2007 Apr;46(4):719-26. doi: 10.1016/j.jhep.2007.01.007. Epub 2007 Jan 29. J Hepatol. 2007. PMID: 17316876 Free PMC article. Review. - Novel cell type-specific antiviral mechanism of interferon gamma action in macrophages.
Presti RM, Popkin DL, Connick M, Paetzold S, Virgin HW 4th. Presti RM, et al. J Exp Med. 2001 Feb 19;193(4):483-96. doi: 10.1084/jem.193.4.483. J Exp Med. 2001. PMID: 11181700 Free PMC article. - Revisiting Hepatitis B Virus: Challenges of Curative Therapies.
Hu J, Protzer U, Siddiqui A. Hu J, et al. J Virol. 2019 Sep 30;93(20):e01032-19. doi: 10.1128/JVI.01032-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31375584 Free PMC article. Review. - La proteins couple use of sequence-specific and non-specific binding modes to engage RNA substrates.
Bayfield MA, Vinayak J, Kerkhofs K, Mansouri-Noori F. Bayfield MA, et al. RNA Biol. 2021 Feb;18(2):168-177. doi: 10.1080/15476286.2019.1582955. Epub 2019 Mar 18. RNA Biol. 2021. PMID: 30777481 Free PMC article. Review. - Control of hepatitis B virus replication by interferons and Toll-like receptor signaling pathways.
Pei RJ, Chen XW, Lu MJ. Pei RJ, et al. World J Gastroenterol. 2014 Sep 7;20(33):11618-29. doi: 10.3748/wjg.v20.i33.11618. World J Gastroenterol. 2014. PMID: 25206268 Free PMC article. Review.
References
- Alonso S, Minty A, Bourlet Y, Buckingham M. Comparison of three actin-coding sequences in the mouse; evolutionary relationships between the actin genes of warm-blooded vertebrates. J Mol Evol. 1986;23:11–22. - PubMed
- Ando K, Guidotti L G, Cerny A, Ishikawa T, Chisari F V. CTL access to tissue antigen is restricted in vivo. J Immunol. 1994;153:482–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials