Selective histocompatibility leukocyte antigen (HLA)-A2 loss caused by aberrant pre-mRNA splicing in 624MEL28 melanoma cells - PubMed (original) (raw)
Selective histocompatibility leukocyte antigen (HLA)-A2 loss caused by aberrant pre-mRNA splicing in 624MEL28 melanoma cells
Z Wang et al. J Exp Med. 1999.
Abstract
Histocompatibility leukocyte antigen (HLA)-A2 is used as a restricting element to present several melanoma-associated antigen (MAA)-derived peptides to cytotoxic T lymphocytes (CTLs). HLA-A2 antigen is selectively lost in primary melanoma lesions and more frequently in metastases. Only scanty information is available about the molecular mechanisms underlying this abnormality, in spite of its potentially negative impact on the clinical course of the disease and on the outcome of T cell-based immunotherapy. Therefore, in this study we have shown that the selective HLA-A2 antigen loss in melanoma cells 624MEL28 is caused by a splicing defect of HLA-A2 pre-mRNA because of a base substitution at the 5' splice donor site of intron 2 of the HLA-A2 gene. As a result, HLA-A2 transcripts are spliced to two aberrant forms, one with exon 2 skipping and the other with intron 2 retention. The latter is not translated because of an early premature stop codon in the retained intron. In contrast, the transcript with exon 2 skipping is translated to a truncated HLA-A2 heavy chain without the alpha(1) domain. Such a polypeptide is synthesized in vitro but is not detectable in cells, probably because of the low steady state level of the corresponding mRNA and the low translation efficiency. These results indicate that a single mutational event in an HLA class I gene is sufficient for loss of the corresponding allele. This may account, at least in part, for the high frequency of selective HLA class I allele loss in melanoma cells. Our conclusion emphasizes the need to implement active specific immunotherapy with a combination of peptides presented by various HLA class I alleles. This strategy may counteract the ability of melanoma cells with selective HLA class I allele loss to escape from immune recognition.
Figures
Figure 1
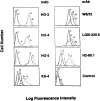
Selective lack of cell surface HLA-A2 antigen expression by 624MEL28 cells. 624MEL28 (a) and 624MEL38 (b) cells were incubated at 37°C for 72 h with IFN-γ (100 U/ml). 624MEL28 (c) and 624MEL38 (d) cells incubated under the same experimental conditions but without IFN-γ were used as controls. At the end of the incubation, cells were harvested, washed with PBS, and incubated with anti–HLA-A2,B17 mAb HO-2, anti–HLA-A2,A28 mAbs HO-4 and HO-5, anti-HLA class I mAb W6/32, anti–HLA-A mAb LGIII-220.6, anti–HLA-B mAb H2-89.1, and anti–HLA-B7 cross-reacting group mAb KS4. After washings with PBS, cells were stained with FITC-GAM. Fluorescence intensity was determined on a FACS® analyzer (Becton Dickinson). Background was determined by incubating cells with FITC-GAM alone (control).
Figure 2
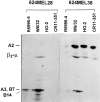
Lack of HLA-A2 heavy chain synthesis by 624MEL28 cells. 624MEL28 and 624MEL38 cells were incubated at 37°C for 72 h with IFN-γ (100 U/ml). Cells were metabolically radiolabeled with [32S]methionine and lysed with 1% NP-40. After indirect immunoprecipitation with anti–β2-μ–free HLA class I heavy chain rabbit serum R5996-4, anti-HLA class I mAb W6/32, anti–HLA-A2,B17 mAb HO-2, and anti–HLA-A2,A28 mAb CR11-351, antigens were analyzed by IEF on a slab gel apparatus. Gels were then processed for fluorography.
Figure 4
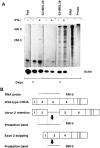
Detection of two alternative forms of HLA-A2 cDNA amplified by RT-PCR from 624MEL28 cells. HLA class I heavy chain cDNA was synthesized from RNA isolated from 624MEL28 cells by reverse transcriptase reaction using HLA class I–specific 3′ primer 3pE3. HLA-A2 cDNA was amplified by PCR using the two pairs of HLA-A2–specific primer (A and B). HLA-A locus cDNA was amplified using an HLA-A locus–specific primer pair (C). The regions from which the primers are derived are indicated below each panel. The corresponding exon regions in the cDNA are indicated by numbers. RT-PCR products were subjected to Southern blotting and hybridized with HLA-A2,A69–specific probe 95V or HLA-A3–specific probe 161D as indicated. DNA amplified from HLA-A2–positive 624MEL38 cells and from HLA-A2–negative Raji cells were used as HLA-A2–positive and –negative controls, respectively. DNA amplified from HLA-A3–negative SKMEL-29.1 cells was used as a negative control for HLA-A3.
Figure 3
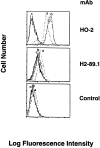
Induction of HLA-A2 antigen expression by 624MEL28 cells after transfection with a wild-type HLA-A2 gene. HLA-A2 gene–transfected 624MEL28 clones 2 (a) and 3 (b) were incubated with anti–HLA-A2,B17 mAb HO-2. After washings with PBS, cells were stained with FITC-GAM. Fluorescence intensity was determined on a FACS® analyzer (Becton Dickinson). Nonspecific fluorescence was determined by incubating cells with FITC-GAM alone. 624MEL28 cells that were not transfected with HLA-A2 gene (c) were used as an HLA-A2–negative control. The anti–HLA-B mAb H2-89.1 was used for comparison.
Figure 6
Comparison of nucleotide and deduced amino acid sequences of the two forms of HLA-A2 cDNA from 624MEL28 cells with those of the wild-type HLA-A2.1 cDNA. The coding region and deduced amino acid sequences of HLA-A2 cDNA corresponding to exon 1, exon 2, and part of exon 3 of the wild-type HLA-A2 cDNA (WT) and of the HLA-A2 cDNA with exon 2 skipping (ST) and intron 2 retention (LT) isolated from 624MEL28 cells are compared. Dashes and blanks indicate sequence identity and deletion, respectively. Intron 2 retention is indicated by a large arrowhead. The stop codon is underlined.
Figure 5
Identification of exon 2 skipping and intron 2 retention in HLA-A2 cDNA from 624MEL28 cells. The 218- and 729-bp RT-PCR products amplified from RNA isolated from 624MEL28 cells using HLA-A2–specific primers 5pE1A2 and 3pE3A2 were purified from agarose gel using a Geneclean II (Bio 101 Inc.) kit. RT-PCR products were then sequenced using a Sequenase 2.0 kit (United States Biochemical Corp.). The autoradiography of the sequencing gels for these RT-PCR products is shown as indicated. The 488-bp RT-PCR products amplified from 624MEL38 cells were sequenced using the same primers as a normal control.
Figure 8
Reduced steady state level of the aberrant HLA-A2 mRNA in 624MEL28 melanoma cells. 624MEL28 cells were incubated at 37°C for 72 h with IFN-γ (100 U/ml). Control cells were incubated under the same experimental conditions, but without IFN-γ. At the end of the incubation, total cellular RNA was extracted from cells and hybridized with a 32P-labeled HLA-A2 riboprobe (496 bases) and with a 32P-labeled human β-actin riboprobe (250 bases). After hybridization, RNA was digested by RNase, and the generated fragments were size fractionated by 5% PAGE. Gels were autoradiographed for 1 or 7 d. RNA from 624MEL38 and Raji cells were used as HLA-A2–positive and –negative controls, respectively. (A) RNA synthesized in vitro by an HLA-A2 genomic fragment containing exon 2, intron 2, and exon 3 (RNA) was used to identify the fragments generated by RNase digestion from the probe protected by the HLA-A2 mRNA containing intron 2. (B) The fragments generated by RNase digestion from the probe protected by a wild-type HLA-A2 mRNA, by HLA-A2 mRNA with exon 2 skipping, and by HLA-A2 mRNA with intron 2 retention are shown diagrammatically.
Figure 7
Identification of a base substitution in intron 2 of the HLA-A2 gene and alternative splicing of HLA-A2 pre-mRNA in 624MEL28 cells. Exon 2 and intron 2 segments of the HLA-A2 gene were amplified by PCR from genomic DNA of 624MEL28 cells using the HLA-A2–specific primer pair 5pE1A2 and 3pE3. HLA-A2 DNA amplified from 624MEL38 cells was used as a control. DNA was sequenced using a Sequenase 2.0 kit. The exon 2–intron 2 boundary and the first two nucleotides of intron 2 are indicated in the DNA sequencing gel (top). The splicing of HLA-A2 pre-mRNA in 624MEL28 cells is compared diagrammatically with that of wild-type HLA-A2 pre-mRNA (bottom).
Figure 9
In vitro synthesis of a truncated HLA class I heavy chain by HLA-A2 mRNA with exon 2 skipping isolated from 624MEL28 cells. The cloned HLA-A2 cDNA lacking exon 2 sequence (E2− A2) was used as a template for mRNA synthesis. The synthesized mRNA was translated in the presence of [35S]methionine in a coupled transcription/translation system driven by T7 promoter (Promega Corp.). A wild-type HLA-A2 cDNA (A2) was used as a control. Translation products were analyzed by SDS-PAGE (A) or were immunoprecipitated by the rabbit anti–β2-μ–free HLA class I heavy chain serum R5996-4 and by the anti–β2-μ–free HLA heavy chain mAb HC-A2 before analysis by SDS-PAGE (B). Normal rabbit serum (NRS) was used as a negative control. The degree of translation was quantitated by scanning densitometry of the intrinsically radiolabeled proteins in the gel (C). The level of RNA synthesis was measured by [32P]UTP incorporation obtained from TCA precipitation (D).
Similar articles
- Quantitative correlation between HLA class I allele expression and recognition of melanoma cells by antigen-specific cytotoxic T lymphocytes.
Rivoltini L, Barracchini KC, Viggiano V, Kawakami Y, Smith A, Mixon A, Restifo NP, Topalian SL, Simonis TB, Rosenberg SA, et al. Rivoltini L, et al. Cancer Res. 1995 Jul 15;55(14):3149-57. Cancer Res. 1995. PMID: 7541714 Free PMC article. - Tumor escape from immune recognition: loss of HLA-A2 melanoma cell surface expression is associated with a complex rearrangement of the short arm of chromosome 6.
Maeurer MJ, Gollin SM, Storkus WJ, Swaney W, Karbach J, Martin D, Castelli C, Salter R, Knuth A, Lotze MT. Maeurer MJ, et al. Clin Cancer Res. 1996 Apr;2(4):641-52. Clin Cancer Res. 1996. PMID: 9816214 - Molecular analysis of the HLA-A2 antigen loss by melanoma cells SK-MEL-29.1.22 and SK-MEL-29.1.29.
Wang Z, Seliger B, Mike N, Momburg F, Knuth A, Ferrone S. Wang Z, et al. Cancer Res. 1998 May 15;58(10):2149-57. Cancer Res. 1998. PMID: 9605759 - Expression of HLA-A2 antigen in human melanoma cell lines and its role in T-cell recognition.
Pandolfi F, Boyle LA, Trentin L, Kurnick JT, Isselbacher KJ, Gattoni-Celli S. Pandolfi F, et al. Cancer Res. 1991 Jun 15;51(12):3164-70. Cancer Res. 1991. PMID: 1904004
Cited by
- Mechanisms of malignant glioma immune resistance and sources of immunosuppression.
Gomez GG, Kruse CA. Gomez GG, et al. Gene Ther Mol Biol. 2006;10(A):133-146. Gene Ther Mol Biol. 2006. PMID: 16810329 Free PMC article. - Epigenetic dysregulation of immune-related pathways in cancer: bioinformatics tools and visualization.
Berglund A, Putney RM, Hamaidi I, Kim S. Berglund A, et al. Exp Mol Med. 2021 May;53(5):761-771. doi: 10.1038/s12276-021-00612-z. Epub 2021 May 7. Exp Mol Med. 2021. PMID: 33963293 Free PMC article. Review. - HLA class I and II genotype of the NCI-60 cell lines.
Adams S, Robbins FM, Chen D, Wagage D, Holbeck SL, Morse HC 3rd, Stroncek D, Marincola FM. Adams S, et al. J Transl Med. 2005 Mar 4;3(1):11. doi: 10.1186/1479-5876-3-11. J Transl Med. 2005. PMID: 15748285 Free PMC article. - Human leukocyte antigen (HLA) class I defects in head and neck cancer: molecular mechanisms and clinical significance.
Ferris RL, Hunt JL, Ferrone S. Ferris RL, et al. Immunol Res. 2005;33(2):113-33. doi: 10.1385/IR:33:2:113. Immunol Res. 2005. PMID: 16234579 Review. - SARS-CoV-2 Selectively Induces the Expression of Unproductive Splicing Isoforms of Interferon, Class I MHC, and Splicing Machinery Genes.
Dias TL, Mamede I, de Toledo NE, Queiroz LR, Castro Í, Polidoro R, Del-Bem LE, Nakaya H, Franco GR. Dias TL, et al. Int J Mol Sci. 2024 May 23;25(11):5671. doi: 10.3390/ijms25115671. Int J Mol Sci. 2024. PMID: 38891862 Free PMC article.
References
- Ferrone S., Marincola F.M. Loss of HLA class I antigens by melanoma cellsmolecular mechanisms, functional significance and clinical relevance. Immunol. Today. 1995;16:487–494. - PubMed
- Kageshita T., Wang Z., Calorini L., Yoshii A., Kimura T., Ono T., Gattoni-Celli S., Ferrone S. Selective loss of human leukocyte class I allospecificities and staining of melanoma cells by monoclonal antibodies recognizing monomorphic determinants of class I human leukocyte antigens. Cancer Res. 1993;53:3349–3354. - PubMed
- Marincola F.M., Shamamian P., Alexander R.B., Gnarra J.R., Turetskaya R.L., Nedospasov S.A., Simonis B., Taubenberger J.K., Yannelli J., Mixon A. Loss of HLA haplotype and B locus down-regulation in melanoma cell lines. J. Immunol. 1994;153:1225–1237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials