Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy - PubMed (original) (raw)
Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy
J C Tardiff et al. J Clin Invest. 1999 Aug.
Abstract
Multiple mutations in cardiac troponin T (cTnT) can cause familial hypertrophic cardiomyopathy (FHC). Patients with cTnT mutations generally exhibit mild or no ventricular hypertrophy, yet demonstrate a high frequency of early sudden death. To understand the functional basis of these phenotypes, we created transgenic mouse lines expressing 30%, 67%, and 92% of their total cTnT as a missense (R92Q) allele analogous to one found in FHC. Similar to a mouse FHC model expressing a truncated cTnT protein, the left ventricles of all R92Q lines are smaller than those of wild-type. In striking contrast to truncation mice, however, the R92Q hearts demonstrate significant induction of atrial natriuretic factor and beta-myosin heavy chain transcripts, interstitial fibrosis, and mitochondrial pathology. Isolated cardiac myocytes from R92Q mice have increased basal sarcomeric activation, impaired relaxation, and shorter sarcomere lengths. Isolated working heart data are consistent, showing hypercontractility and diastolic dysfunction, both of which are common findings in patients with FHC. These mice represent the first disease model to exhibit hypercontractility, as well as a unique model system for exploring the cellular pathogenesis of FHC. The distinct phenotypes of mice with different TnT alleles suggest that the clinical heterogeneity of FHC is at least partially due to allele-specific mechanisms.
Figures
Figure 1
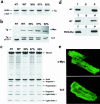
R92Q-TnT and truncation-TnT map to 2 distinct functional domains. (a) Exon composition of human cTnT, with positions of disease alleles Arg92Gln and intron 15 G→A marked above. The representative murine transgene allele is in parenthesis below. Tr-Myc represents a loss of exons 15 and 16. Functional domains are depicted by filled bars, with associated protein binding sites below. Location of translational start and stop codons are represented by ATG and TAG, respectively. (b) Constructs used to generate wild-type (WT) and Arg92Gln (R92Q) Myc-tagged murine cardiac TnT.
Figure 2
Expression of WT-Myc and R92Q-Myc proteins in cardiac tissue. (a) Coomassie-stained SDS-PAGE gel of myofibrils isolated from non-Tg (NT), WT-Myc (WT), and 3 independent R92Q-Myc lines. A total of 5 μg myofibril protein was loaded per lane. Addition of the c_-myc_ epitope tag decreases the mobility of the Tg protein (Tg) and allows unambiguous identification. Percentages (30%, 67%, 92%) represent Tg/endogenous cTnT ratios for the 3 R92Q-Myc lines. (b) Western blot analysis of myofibrils subjected to the same SDS-PAGE conditions as noted in a. Identical blots were probed with either a c_-myc_ or cTnT mAb as indicated. The cTnT MAb detects both Tg and endogenous cTnT, and their relative positions are marked by arrows. The additional band immediately above the cTnT band (seen best in the NT lane) represents a known murine cTnT isoform. Note the progressive decrease in endogenous cTnT protein amounts among the 3 R92Q-Myc lines. (c) Myofibrils were purified from mouse hearts, subjected to SDS-PAGE, and Coomassie stained. Myofibrillar stoichiometry is maintained in all Tg lines. (d) Fractionation of cTnT. Three separate fractions were analyzed (T = total, S = supernatant, and P = pellet). Immunoblots of fractions from non-Tg (NT), WT-Myc (WT), and R92Q-67% (R92Q-Myc) were loaded for equal signal intensity and probed with either a cTnT (non-Tg) or c_-myc_ (WT-Myc and R92Q-Myc) mAb as indicated. No Tg protein was detected in the S fraction for either WT-Myc or R92Q-Myc. (e) Transgene protein incorporation. Shown are confocal images of isolated adult cardiac myocytes probed with either c-myc (top; R92Q-67%) or TnT (bottom; non-Tg) mAb. ×4,300.
Figure 3
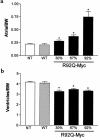
Heterozygous R92Q-Myc mice demonstrate decreased ventricular mass and dose-dependent changes in atrial mass. (a) Atrial weight/body weight ratios (milligram of chamber weight per gram of body weight [BW]) in 4- to 6-month-old animals. Error bars represent SEM. P values are vs. non-Tg (NT) animals (1-way ANOVA). (b) Ventricular weight/body weight ratios from the same mice as shown in a. *P < 0.0001.
Figure 4
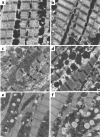
R92Q-Myc mice demonstrate myocellular disarray and fibrosis. Cardiac sections from 4- to 5-month-old non-Tg (a), R92Q-Myc-67% (b and c), WT-Myc (d), and R92Q-Myc-92% (e and f). a, b, d, and e were stained with hematoxylin and eosin. Note markedly pleiotropic nuclei and myocellular degeneration in b and e. e also demonstrates clear myocellular disarray and inflammation. c and f are stained with Masson’s trichrome and demonstrate dose-dependent myocardial fibrosis. All panels ×400.
Figure 5
Ultrastructural abnormalities in cTnT-related FHC mice are allele specific. Representative electron micrographs from left ventricular tissue isolated from non-Tg (a), WT-Myc (b), truncation-Myc (c and d), and R92Q-Myc (e and f) mice at 5 weeks of age. Sections from non-Tg and WT-Myc both demonstrate ordered arrays of myofibrils and normal mitochondrial morphology and number. Sections from 2 independent truncation-Myc lines reveal frequent misregistration of Z bands, myofibrillar disarray, and degeneration. Mitochondria are variable in size and morphology is normal. In comparison, both the R92Q-Myc lines (e = 67%, f = 92%) demonstrate largely preserved sarcomeric structure; however, note the lipid deposition and increased numbers of small mitochondria with loss of well-defined membranes and cristae. ×15,000. Scale bar: 1 μm.
Figure 6
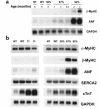
Induction of hypertrophic markers in R92Q-Myc mice. (a) Induction time-course Northern blot analysis of total ventricular RNA isolated from non-Tg (NT), WT-Myc (WT), and all 3 R92Q-Myc Tg lines. Age at time of RNA isolation is noted for each lane. The blot was serially hybridized with a radiolabeled β-MyHC oligonucleotide probe and ANF and GAPDH cDNA probes as described in Methods. (b) Northern blot analysis of total ventricular RNA isolated from non-Tg (NT), WT-Myc (WT), truncation-Myc (Tr), and all 3 R92Q-Myc Tg lines. A total of 10 μg of total RNA was loaded per lane. The blot was serially hybridized with α-MyHC and β-MyHC oligonucleotide probes, followed by ANF, SERCA2, cTnT, and GAPDH cDNA probes. Note that the cTnT probe detects both endogenous (as seen in NT lanes) and transgene transcripts.
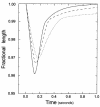
Figure 7
Representative ventricular myocyte-shortening dynamics. Non-Tg (solid line), WT-Myc (dashed line), and R92Q-Myc (dotted line) are shown above. “Fractional length” (ordinate label) refers to cell shortening as a fraction of resting cell length. Myocyte-shortening dynamics for each cell type were reconstructed from the data presented in Table 1 (and mean times to 25%, 75%, and 90% relengthening; not reported in Table 1) by interpolation.
Similar articles
- A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy.
Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, Schwartz S, Robbins J, Leinwand LA. Tardiff JC, et al. J Clin Invest. 1998 Jun 15;101(12):2800-11. doi: 10.1172/JCI2389. J Clin Invest. 1998. PMID: 9637714 Free PMC article. - Cardiac myosin heavy chain isoform exchange alters the phenotype of cTnT-related cardiomyopathies in mouse hearts.
Rice R, Guinto P, Dowell-Martino C, He H, Hoyer K, Krenz M, Robbins J, Ingwall JS, Tardiff JC. Rice R, et al. J Mol Cell Cardiol. 2010 May;48(5):979-88. doi: 10.1016/j.yjmcc.2009.11.018. Epub 2009 Dec 31. J Mol Cell Cardiol. 2010. PMID: 20004663 Free PMC article. - Pathogenesis of Hypertrophic Cardiomyopathy is Mutation Rather Than Disease Specific: A Comparison of the Cardiac Troponin T E163R and R92Q Mouse Models.
Ferrantini C, Coppini R, Pioner JM, Gentile F, Tosi B, Mazzoni L, Scellini B, Piroddi N, Laurino A, Santini L, Spinelli V, Sacconi L, De Tombe P, Moore R, Tardiff J, Mugelli A, Olivotto I, Cerbai E, Tesi C, Poggesi C. Ferrantini C, et al. J Am Heart Assoc. 2017 Jul 22;6(7):e005407. doi: 10.1161/JAHA.116.005407. J Am Heart Assoc. 2017. PMID: 28735292 Free PMC article. - Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes.
Tardiff JC. Tardiff JC. Heart Fail Rev. 2005 Sep;10(3):237-48. doi: 10.1007/s10741-005-5253-5. Heart Fail Rev. 2005. PMID: 16416046 Review. - [Genotype-phenotype correlations in familial hypertrophic cardiomyopathy].
Anan R, Niimura H, Tei C. Anan R, et al. Nihon Rinsho. 2000 Jan;58(1):134-40. Nihon Rinsho. 2000. PMID: 10885301 Review. Japanese.
Cited by
- Allele-specific dysregulation of lipid and energy metabolism in early-stage hypertrophic cardiomyopathy.
Vaniya A, Karlstaedt A, Gulkok D, Thottakara T, Liu Y, Fan S, Eades H, Vakrou S, Fukunaga R, Vernon HJ, Fiehn O, Roselle Abraham M. Vaniya A, et al. J Mol Cell Cardiol Plus. 2024 Jun;8:100073. doi: 10.1016/j.jmccpl.2024.100073. Epub 2024 Mar 31. J Mol Cell Cardiol Plus. 2024. PMID: 39430912 Free PMC article. - Effects of R92 mutations in mouse cardiac troponin T are influenced by changes in myosin heavy chain isoform.
Ford SJ, Mamidi R, Jimenez J, Tardiff JC, Chandra M. Ford SJ, et al. J Mol Cell Cardiol. 2012 Oct;53(4):542-51. doi: 10.1016/j.yjmcc.2012.07.018. Epub 2012 Aug 4. J Mol Cell Cardiol. 2012. PMID: 22884844 Free PMC article. - Sustained activation of Akt elicits mitochondrial dysfunction to block Plasmodium falciparum infection in the mosquito host.
Luckhart S, Giulivi C, Drexler AL, Antonova-Koch Y, Sakaguchi D, Napoli E, Wong S, Price MS, Eigenheer R, Phinney BS, Pakpour N, Pietri JE, Cheung K, Georgis M, Riehle M. Luckhart S, et al. PLoS Pathog. 2013 Feb;9(2):e1003180. doi: 10.1371/journal.ppat.1003180. Epub 2013 Feb 28. PLoS Pathog. 2013. PMID: 23468624 Free PMC article. - Narrative review: harnessing molecular genetics for the diagnosis and management of hypertrophic cardiomyopathy.
Wang L, Seidman JG, Seidman CE. Wang L, et al. Ann Intern Med. 2010 Apr 20;152(8):513-20, W181. doi: 10.7326/0003-4819-152-8-201004200-00008. Ann Intern Med. 2010. PMID: 20404382 Free PMC article. Review. - Rescue of familial cardiomyopathies by modifications at the level of sarcomere and Ca2+ fluxes.
Alves ML, Gaffin RD, Wolska BM. Alves ML, et al. J Mol Cell Cardiol. 2010 May;48(5):834-42. doi: 10.1016/j.yjmcc.2010.01.003. Epub 2010 Jan 15. J Mol Cell Cardiol. 2010. PMID: 20079744 Free PMC article. Review.
References
- Towbin J. The role of cytoskeletal proteins in cardiomyopathies. Curr Opin Cell Biol. 1998;10:131–139. - PubMed
- Watkins H, Seidman JG, Seidman CE. Familial hypertrophic cardiomyopathy: a genetic model of cardiac hypertrophy. Hum Mol Genet. 1995;4:1721–1727. - PubMed
- Sweeney HL, Straceski AJ, Leinwand LA, Faust L. Heterologous expression of a cardiomyopathic myosin that is defective in its actin interaction. J Biol Chem. 1994;269:1603–1605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL-060546/HL/NHLBI NIH HHS/United States
- HL-50560/HL/NHLBI NIH HHS/United States
- R37 HL050560/HL/NHLBI NIH HHS/United States
- R01 HL040306/HL/NHLBI NIH HHS/United States
- HL-40306/HL/NHLBI NIH HHS/United States
- R01 HL050560/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials