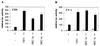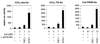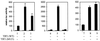The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription - PubMed (original) (raw)
The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription
P Alen et al. Mol Cell Biol. 1999 Sep.
Abstract
Steroid receptors are conditional transcription factors that, upon binding to their response elements, regulate the expression of target genes via direct protein interactions with transcriptional coactivators. We have analyzed the functional interactions between the androgen receptor (AR) and 160-kDa nuclear receptor coactivators. Upon overexpression in mammalian cells, these coactivators enhance the transcriptional activity of both the amino-terminal domain (NTD) and the ligand-binding domain (LBD) of the AR. The coactivator activity for the LBD is strictly ligand-controlled and depends on the nature of the DNA-binding domain to which it is fused. We demonstrate that the NTD physically interacts with coactivators and with the LBD and that this interaction, like the functional interaction between the LBD and p160 coactivators, relies on the activation function 2 (AF2) core domain. The mutation of a highly conserved lysine residue in the predicted helix 3 of the LBD (K720A), however, blunts the functional interaction with coactivators but not with the NTD. Moreover, this mutation does not affect the transcriptional activity of the full-size AR. A mutation in the NTD of activation function AF1a (I182A/L183A), which dramatically impairs the activity of the AR, has no effect on the intrinsic transcriptional activity of the NTD but interferes with the cooperation between the NTD and the LBD. Finally, p160 proteins in which the three LXXLL motifs are mutated retain most of their coactivator activity for the full-size AR, although they are no longer functional for the isolated LBD. Together, these data suggest that in the native AR the efficient recruitment of coactivators requires a functional association of the NTD with the LBD and that the binding of coactivators occurs primarily through the NTD.
Figures
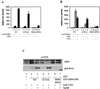
FIG. 1
p160 coactivators stimulate the transcriptional activity of the full-size hAR. COS 7 cells (A) and CV-1 cells (B) were cotransfected with pMMTV-luc (500 ng) and pSG5-hAR (50 ng), along with 500 ng of either empty pSG5 (bar 1) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (bar 2 to 4). The cells were incubated with DCC-treated medium, either supplemented (solid bars) or not (open bars) with 1 nM R1881 24 h before being harvested. The luciferase activities were corrected for transfection efficiency by using the β-galactosidase activities. The data are presented as relative activities (± SEM). The luciferase activity measured in the presence of ligand but without cotransfected coactivator was arbitrarily set to 100, and test values were calculated accordingly.
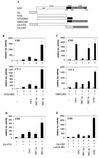
FIG. 2
The hAR AF1 and AF2 functions are both stimulated by p160 coactivators. (A) Schematic representation of the hAR and the constructs used in this study. Depicted are the NTD, the centrally located DBD, the hinge region (hatched box), and the C-terminal LBD of the hAR, and the DBD of the yeast transcription factor GAL4 (G4). The numbers refer to amino acid positions. ∗∗, position of the residues mutated in the AF1a mutant (I182A/L183A); ◊, position of the H3 mutant (K720A); ▵▵, position of the AF2 mutant (I898A/I899A). (B) COS cells (upper graph) and CV-1 cells (lower graph) were transfected with pMMTV-luc (250 ng) without (bars 1) or with (bars 2 to 5) an expression vector encoding NTD-DBD (125 ng) and with either empty pSG5 (bars 1 and 2) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (900 ng). After 24 h, the cells were harvested and luciferase and β-galactosidase activities were measured as described in Materials and Methods. The activity of NTD-DBD in the absence of exogenously added coactivator was given the value 100. All other activities were calculated accordingly. (C) COS cells (upper graph) and CV-1 cells (lower graph) were cotransfected with pMMTV-luc (250 ng), pSG5-DBD/LBD (25 ng), and either empty pSG5 (bar 1) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (250 ng) (bars 2 to 5) and incubated for 24 h in DCC-treated medium, either supplemented (solid bars) or not (open bars) with 1 nM R1881. They were harvested and processed as described in Materials and Methods. (D) COS 7 cells were transfected with the p(G4)5-tata-luc reporter (250 ng) without (bar 1) or with (bars 2 to 5) an expression vector for G4-NTD (25 ng) and with either empty pSG5 (bars 1 and 2) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (bars 3 to 5; 250 ng) and treated as described for panel B. (E) COS 7 cells were transfected as for Fig. 2C, except that the p(G4)5-tata-luc reporter vector was used and pSG5-DBD/LBD was replaced by an expression vector encoding G4-LBD. The cells were treated with 1 nM R1881 as indicated. The luciferase activity measured in the presence of R1881 but without ectopically expressed coactivator (bar 2) was set to 100, and test values were calculated accordingly. The error bars indicate the SEM.
FIG. 3
hAR NTD efficiently stimulates the transcriptional activity of G4-LBD, independent of the promoter context. COS 7 cells were cotransfected with either p(G4)5-tata-luc (left graph), p(G4)5-TK-luc (center graph), or pGal-50hI16-luc (right graph), along with pG4-LBD and either empty pSG5 (bars 1, 2, 5, 6, 9, and 10), pSG5-TIF2 (bars 3, 7, and 11), or pSG5-NTD (bars 4, 8, and 12), as for Fig. 2E. The cells were incubated with DCC-treated medium, either supplemented (+) or not (−) with 1 nM R1881 for 24 h and subsequently processed as described in Materials and Methods. In each case, the luciferase activity measured in the presence of hormone but in the absence of exogenously added coactivator was arbitrarily set to 100, and the values for the other conditions were calculated accordingly. The error bars indicate the SEM.
FIG. 4
hAR NTD physically interacts with SRC-1e and with the hAR LBD in COS cells. (A) COS cells were transfected with expression vectors for either GST alone or a fusion protein of GST with the hAR NTD, along with an expression vector for FLAG-tagged SRC-1e. After 24 h of incubation at 37°C, extracts were prepared and protein complexes bound to GST or GST-NTD were precipitated with glutathione Sepharose beads, immunoblotted, and stained with monoclonal anti-FLAG antibody. (B) COS cells were transfected and treated as described for panel A, except that GST fusion proteins of the DBD-LBD regions of the hAR and the mER were used. The cells were treated without hormone (lanes 1, 2, and 4) or with 100 nM of either R1881 (lane 3) or estradiol (lane 5) 24 h prior to the preparation of protein extracts and GST pull down. (C) COS cells were transfected and treated as for panel B, except that an expression vector for the hAR NTD was used instead of FLAG-tagged SRC-1e. The immunoblots were probed with a polyclonal anti-AR antiserum. +, present; −, absent.
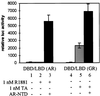
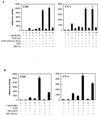
FIG. 5
Coexpression of the hAR NTD strongly stimulates the transcriptional activity of the hAR LBD but has only weak effects on the rGR AF2 function. COS 7 cells were transfected with 250 ng of pMMTV-luc, along with 25 ng of either pSG5-DBD/LBD(AR) or pSG5-DBD/LBD(GR) and 250 ng of either empty pSG5 (bar 1, 2, 4, and 5) or pSG5-NTD (bar 3 and 6), and treated for 24 h without hormone (bar 1 and 4), with 1 nM R1881 (bars 2 and 3), or with 1 nM triamcinolone acetonide (TA) (bars 5 and 6). Luciferase and β-galactosidase activities were measured as described in Materials and Methods. The luciferase activity measured with DBD-LBD(AR) in the presence of R1881 and in the absence of NTD (bar 2) was arbitrarily set to 100. The activities of all other conditions were calculated relative to this standard. The error bars indicate the SEM.
FIG. 6
The effect of the K720A and I898A/I899A mutations on the transcriptional activities of the native hAR and on the DBD-LBD construct. (A) COS 7 cells were cotransfected with 250 ng of pMMTV-luc and 25 ng of expression vector for either the full-size wild-type hAR, the K720A mutant, or the I898A/I899A mutant, along with 250 ng of either empty pSG5 (bars 1, 2, 5, 6, 9, and 10) or pSG5-TIF2 (bars 3, 4, 7, 8, 11, and 12) and incubated for 24 h with DCC-treated medium without (open bars) or with (solid bars) 1 nM R1881. The measured luciferase activities were corrected for protein content and β-galactosidase activity and are expressed as relative values, with the activity of the native receptor in the presence of R1881 taken as 100. (B) COS 7 cells were transfected as described for panel A, but instead of expression vectors for full-size receptors, 25 ng of the plasmids encoding the DBD-LBD fragments of either the wild-type receptor, the K720A or the I898A/I899A mutants were used, along with 250 ng of either empty pSG5 (bars 1, 2, 5, 6, 9, and 10), pSG5-TIF2 (bars 3, 7, and 11) or pSG5-NTD (bars 4, 8, and 12). The cells were either left untreated (bars 1, 5, and 6) or treated with 1 nM R1881 (bars 2 to 4, 6 to 8, and 10 to 12) for 24 h before being harvested. The luciferase activity measured for the wild-type DBD-LBD construct in the presence of hormone was taken as 100, and test values were calculated. (C) GST pull-down experiments were performed as described in Materials and Methods and in the legend to Fig. 4. The immunoblots were probed with an anti-AR antiserum. +, present; −, absent. The error bars indicate the SEM.
FIG. 7
The effect of mutation of the three LXXLL motifs in TIF2 on the transcriptional activity of the hAR. COS-7 cells were transfected with 250 ng of pMMTV-luc reporter and 25 ng of pSG5-hAR (left graph), 25 ng of pSG5-DBD/LBD (center graph), or 125 ng of NTD-DBD expression vector (right graph), along with either 250 ng (bars 1 and 4) or 900 ng (bar 7) of empty pSG5, 250 ng (bars 2 and 5) or 900 ng (bar 8) of pSG5-TIF2(WT), or 250 ng (bars 3 and 6) or 900 ng (bar 9) of pSG5-TIF2(M123). The cells transfected with NTD-DBD (right graph) were incubated for 24 h in DCC-treated medium, and those transfected with the native hAR or DBD-LBD (left and center graphs) received DCC-treated medium supplemented with 1 nM R1881 (solid bars) or vehicle (ethanol) alone (open bars). The luciferase activities were corrected for transfection efficiency by using the β-galactosidase activities. +, present; −, absent. The error bars indicate the SEM.
FIG. 8
Effect of mutation of the AF1a function on the functional NTD-LBD interaction. (A) COS 7 cells were cotransfected with 250 ng of pMMTV-luc and 25 ng of expression vector for the wild-type hAR or the I182A/L183A mutant, along with 250 ng of either empty pSG5 (bars 1 and 3) or pSG5-TIF2 (bars 2 and 4) and incubated with DCC-treated medium, supplemented with vehicle alone (ethanol) (open bars) or 1 nM R1881 (solid bars). The measured luciferase activities were corrected for β-galactosidase activity and are expressed as relative values, with the activity of the wild-type receptor in the presence of hormone but without coexpression of TIF2 set to 100. (B) COS cells were transfected with pMMTV-luc (250 ng), expression vectors for either the wild-type (bars 1 and 3) or the I182A/L183A mutated (bars 2 and 4) NTD-DBD constructs and either empty pSG5 (bars 1 and 2) or pSG5-SRC-1e (bars 3 and 4). Extracts were prepared, and luciferase activities and β-galactosidase activities were measured as described in Materials and Methods. (C) COS 7 cells were transfected with 250 ng of pMMTV-luc and 25 ng of expression plasmid for the wild-type DBD-LBD fragment, along with 250 ng of either empty pSG5 (bars 1 and 2) or expression vector for the wild-type hAR NTD (bar 3) or the NTD carrying the I182A/L183A mutation (bar 4). The cells were treated for 24 h without (bar 1) or with (bars 2 to 4) 1 nM R1881. The luciferase activities are presented relative to the activity measured in the presence of hormone but without cotransfected NTD. (D) GST pull-down experiments were carried out as for Fig. 4C with either GST alone or a GST–DBD-LBD fusion protein and the wild-type (lanes 1 to 3) or I182A/L183A mutated (lane 4) hAR NTD. (E) COS 7 cells were transfected as for panel C, except that plasmids directing the expression of the NTD fused to the VP16 activating domain were used. The luciferase activity measured in the presence of ligand but in the absence of VP16 fusion protein was taken as 100. Test values were calculated accordingly. +, present; −, absent. The error bars indicate the SEM.
FIG. 9
hAR NTD and TIF2 cooperatively stimulate the transcriptional activity of the hAR LBD. (A) COS-7 cells (left graph) or CV-1 cells (right graph) were cotransfected with 250 ng of pMMTV-luc, 25 ng of expression vector for the DBD-LBD construct, and 250 ng of expression vector for either the wild-type or I182A/L183A mutated NTD, for TIF2, or for SRC-1e as indicated. Under the conditions where only one cofactor was transfected (bars 1 to 6), empty pSG5 plasmid was added to keep the total amount of transfected DNA constant. The cells were incubated for 24 h without (bars 1) or with (bars 2 to 10) 1 nM R1881 before harvesting and measurement of luciferase and β-galactosidase activities. The luciferase activity measured in the presence of hormone, but without cotransfected NTD or coactivator (bars 2), was set to 100, and test values were calculated accordingly. (B) COS-7 cells (left graph) or CV-1 cells (right graph) were cotransfected with 250 ng of pMMTV-luc, 25 ng of expression vector for the DBD-LBD construct, and 250 ng of expression vector for the hAR NTD, TIF2(WT), or TIF2(M123), as indicated. For bars 1 to 4 and bars 6, empty pSG5 was added to keep the total amount of transfected DNA constant. The cells were further processed as described for panel A. +, present; −, absent. The error bars indicate the SEM.
Similar articles
- Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins.
Ma H, Hong H, Huang SM, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Ma H, et al. Mol Cell Biol. 1999 Sep;19(9):6164-73. doi: 10.1128/MCB.19.9.6164. Mol Cell Biol. 1999. PMID: 10454563 Free PMC article. - Beta-catenin binds to the activation function 2 region of the androgen receptor and modulates the effects of the N-terminal domain and TIF2 on ligand-dependent transcription.
Song LN, Herrell R, Byers S, Shah S, Wilson EM, Gelmann EP. Song LN, et al. Mol Cell Biol. 2003 Mar;23(5):1674-87. doi: 10.1128/MCB.23.5.1674-1687.2003. Mol Cell Biol. 2003. PMID: 12588987 Free PMC article. - Structural features discriminate androgen receptor N/C terminal and coactivator interactions.
Askew EB, Minges JT, Hnat AT, Wilson EM. Askew EB, et al. Mol Cell Endocrinol. 2012 Jan 30;348(2):403-10. doi: 10.1016/j.mce.2011.03.026. Epub 2011 Jun 1. Mol Cell Endocrinol. 2012. PMID: 21664945 Free PMC article. Review. - The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors.
Edwards DP. Edwards DP. J Mammary Gland Biol Neoplasia. 2000 Jul;5(3):307-24. doi: 10.1023/a:1009503029176. J Mammary Gland Biol Neoplasia. 2000. PMID: 14973393 Review.
Cited by
- Nuclear compartmentalization of N-CoR and its interactions with steroid receptors.
Wu Y, Kawate H, Ohnaka K, Nawata H, Takayanagi R. Wu Y, et al. Mol Cell Biol. 2006 Sep;26(17):6633-55. doi: 10.1128/MCB.01534-05. Mol Cell Biol. 2006. PMID: 16914745 Free PMC article. - Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2.
Benecke A, Chambon P, Gronemeyer H. Benecke A, et al. EMBO Rep. 2000 Aug;1(2):151-7. doi: 10.1093/embo-reports/kvd028. EMBO Rep. 2000. PMID: 11265755 Free PMC article. - Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function.
Fu M, Wang C, Wang J, Zhang X, Sakamaki T, Yeung YG, Chang C, Hopp T, Fuqua SA, Jaffray E, Hay RT, Palvimo JJ, Jänne OA, Pestell RG. Fu M, et al. Mol Cell Biol. 2002 May;22(10):3373-88. doi: 10.1128/MCB.22.10.3373-3388.2002. Mol Cell Biol. 2002. PMID: 11971970 Free PMC article. - Regulation of androgen receptor-dependent transcription by coactivator MED1 is mediated through a newly discovered noncanonical binding motif.
Jin F, Claessens F, Fondell JD. Jin F, et al. J Biol Chem. 2012 Jan 6;287(2):858-70. doi: 10.1074/jbc.M111.304519. Epub 2011 Nov 18. J Biol Chem. 2012. PMID: 22102282 Free PMC article. - The p160 coactivator PAS-B motif stabilizes nuclear receptor binding and contributes to isoform-specific regulation by thyroid hormone receptors.
Privalsky ML, Lee S, Hahm JB, Young BM, Fong RN, Chan IH. Privalsky ML, et al. J Biol Chem. 2009 Jul 17;284(29):19554-63. doi: 10.1074/jbc.M109.007542. Epub 2009 Jun 1. J Biol Chem. 2009. PMID: 19487700 Free PMC article.
References
- Alen P, Claessens F, Schoenmakers E, Swinnen J V, Verhoeven G, Rombauts W, Peeters B. Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1α with multiple steroid receptors and identification of an internally deleted ELE1β isoform. Mol Endocrinol. 1999;13:117–128. - PubMed
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous