hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage - PubMed (original) (raw)
hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage
D R Duckett et al. Proc Natl Acad Sci U S A. 1999.
Abstract
hMSH2.hMSH6 heterodimer (hMutSalpha) and hMLH1.hPMS2 complex (hMutLalpha) have been implicated in the cytotoxic response of mammalian cells to a number of DNA-damaging compounds, including methylating agents that produce O(6)-methylguanine (O(6)MeG) adducts. This study demonstrates that O(6)MeG lesions, in which the damaged base is paired with either T or C, are subject to excision repair in a reaction that depends on a functional mismatch repair system. Furthermore, treatment of human cells with the S(N)1 DNA methylators N-methyl-N-nitrosourea or N-methyl-N'-nitro-N-nitrosoguanidine results in p53 phosphorylation on serine residues 15 and 392, and these phosphorylation events depend on the presence of functional hMutSalpha and hMutLalpha. Coupled with the previous demonstration that O(6)MeG.T and O(6)MeG.C pairs are recognized by hMutSalpha, these results implicate action of the mismatch repair system in the initial step of a damage-signaling cascade that can lead to cell-cycle checkpoint activation or cell death in response to DNA methylator damage.
Figures
Figure 1
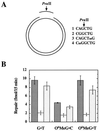
O6MeG lesions are subject to processing by the human mismatch repair system. (A) Oligonucleotides (51 mer) containing _Pvu_II sequence variants shown (mG indicates O6MeG) were hybridized to a circular 6,490-bp DNA containing a 51-nt gap in the complementary DNA strand (Experimental Procedures) to yield the four substrates used in this study. Hybridization of oligonucleotide 1 yields a homoduplex with a normal _Pvu_II recognition sequence, whereas oligonucleotides 2, 3, or 4 generate substrates containing a G⋅T mismatch, an O6MeG⋅C bp, or an O6MeG⋅T lesion, respectively. Presence of the mismatch or O6MeG within the _Pvu_II recognition site renders the DNA resistant to hydrolysis by this enzyme, with sensitivity restored on repair. (B) Mismatch repair assays (Experimental Procedures) scored repair on the incised strand that restored an intact _Pvu_II recognition sequence. Dark grey bars, TK6 nuclear extract; white bars, MT1 nuclear extract; medium grey bars, hMutSα-supplemented MT1 nuclear extract. Values shown are the average of four determinations ± one standard deviation. Because TK6 and MT1 cells are deficient in the O6-alkylguanine-DNA alkyltransferase, complications caused by repair by this activity are obviated.
Figure 2
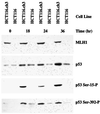
MNU-induced Ser-15 phosphorylation of p53 depends on MSH6 function, but that induced by etoposide does not. TK6 or MT1 cells were treated with 10 μM MNU (A) or 8 μM etoposide (B) for the indicated times, and extract (30 μg) subjected to SDS gel electrophoresis and Western blotting by using antibodies against hMLH1, p53, or affinity-purified antibody specific for the Ser-15 phosphorylated form of p53 (Experimental Procedures). The left two lanes in each p53 blot correspond to 2–5 ng of purified recombinant human p53 protein produced in Escherichia coli that was either untreated or phosphorylated in vitro by using near-homogeneous DNA protein kinase and Ku protein. This kinase is known to phosphorylate p53 on Ser residues 15 and 37 (58).
Figure 3
Phosphorylation of p53 at Ser-392 is induced by methylnitrosourea in a _MSH6_-dependent manner. TK6 or MT1 cells were treated with 10 μM MNU for the indicated time (A) or exposed to 20 J/m2 of 254 nm UV light and then incubated in complete medium for the times indicated (B). Samples of extract (30 μg) were analyzed by SDS gel electrophoresis and immunological blot as described in Fig. 2, except that phosphopeptide-specific antibody was directed against the phosphorylated form of Ser-392.
Figure 4
Phosphorylation of p53 at Ser-15 and Ser-392 is induced by _N_-methyl-_N′_-nitro-_N_-nitrosoguanidine in a _MLH1_-dependent manner. HCT116 or HCT116.ch3 were treated with 5 μM _N_-methyl-N ′-nitro-_N_-nitrosoguanidine (Experimental Procedures) and then incubated in complete medium for the indicated time. Samples of extract (30 μg) were analyzed by SDS gel electrophoresis and immunological blot by using the antisera indicated.
References
- Branch P, Aquilina G, Bignami M, Karran P. Nature (London) 1993;362:652–654. - PubMed
- Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Cancer Res. 1994;54:4308–4312. - PubMed
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. - PubMed
- Drummond J T, Anthoney A, Brown R, Modrich P. J Biol Chem. 1996;271:19645–19648. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous