Recruitment of Nanos to hunchback mRNA by Pumilio - PubMed (original) (raw)
Recruitment of Nanos to hunchback mRNA by Pumilio
J Sonoda et al. Genes Dev. 1999.
Abstract
Translational regulation of hunchback (hb) mRNA is essential for posterior patterning of the Drosophila embryo. This regulation is mediated by sequences in the 3'-untranslated region of hb mRNA (the Nanos response elements or NREs), as well as two trans-acting factors-Nanos and Pumilio. Pumilio recognizes the NREs via a conserved binding motif. The mechanism of Nanos action has not been clear. In this report we use protein-protein and protein-RNA interaction assays in yeast and in vitro to show that Nanos forms a ternary complex with the RNA-binding domain of Pumilio and the NRE. Mutant forms of the NRE, Nos, and Pum that do not regulate hb mRNA normally in embryos do not assemble normally into a ternary complex. In particular, recruitment of Nos is dependent on bases in the center of the NRE, on the carboxy-terminal Cys/His domain of Nos, and on residues in the eighth repeat of the Pum RNA-binding domain. These residues differ in a closely related human protein that also binds to the NRE but cannot recruit Drosophila Nos. Taken together, these findings suggest models for how Nos and Pum collaboratively target hb mRNA. More generally, they suggest that Pum-like proteins from other species may also act by recruiting cofactors to regulate translation.
Figures
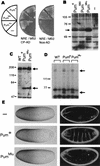
Figure 1
A Pum/NRE/Nos ternary complex in yeast. Experimental plans (A,C) and results (B,D). In B, yeast strains were transformed with plasmids that direct the synthesis of either hybrid molecules (i.e., Pum fused to GAL4–DBD) or empty vectors (i.e., DBD). Formation of the ternary complex (A) activates transcription of HIS3, allowing growth in the absence of exogenous His (B). In C and D, a Nos–AD fusion does not bind to the NRE in the absence of Pum, and HIS3 transcription is not stimulated. As a control, the Pum–AD fusion binds to NRE+ but not NRE21 in this assay (Fig. 2).
Figure 2
Bases in the center of the NRE are required to recruit Nos. (A) Sequence of NRE+ and mutants that reduce its activity in embryos (Murata and Wharton 1995; Wharton et al. 1998). Bases conserved among NREs in bcd and hb genes from various Drosophila species are boxed. Mutations above reduce binding of Pum and those below prevent incorporation of Nos into a ternary complex. Mutant NREs used in these studies, each bearing a dinucleotide substitution, are named according to their position. (B) In vivo yeast assays of Pum/NRE/Nos ternary complex formation. Yeast expressing the Nos, Pum, or CP derivatives indicated were also transformed with plasmids that direct the synthesis of RNAs that contain either the MS2 hairpin (the CP-binding site), or the NRE, or both, as indicated in the drawing at left. The top plate monitors incorporation of Nos into a ternary complex, as shown schematically in Fig. 1A; the bottom plate monitors binding of Pum, as shown schematically in the drawing at right. (C) Northern blot analysis to assay the level of RNA accumulation in yeast. Samples of RNA from yeast transformed either with empty vector (−) or plasmids that direct synthesis of each of the indicated chimeric RNAs were probed with NRE sequences. Each lane contains approximately the same amount of low-molecular-weight RNA as determined by staining with ethidium bromide. Note that the NRE+ RNA (i.e., lacking the MS2 hairpin) accumulates to a higher level than do the NRE/MS2 chimeric RNAs. Compared with yeast expressing the NRE+/MS2 chimera (also cotransformed with Pum–DBD and Nos–AD plasmids), yeast expressing the NRE+ RNA grow on His− media in the presence of higher levels of the HIS3 competitor 3-aminotriazole and express higher levels of LacZ (not shown). These observations suggest that the concentration of RNA substrate is limiting in the yeast ternary complex assay.
Figure 2
Bases in the center of the NRE are required to recruit Nos. (A) Sequence of NRE+ and mutants that reduce its activity in embryos (Murata and Wharton 1995; Wharton et al. 1998). Bases conserved among NREs in bcd and hb genes from various Drosophila species are boxed. Mutations above reduce binding of Pum and those below prevent incorporation of Nos into a ternary complex. Mutant NREs used in these studies, each bearing a dinucleotide substitution, are named according to their position. (B) In vivo yeast assays of Pum/NRE/Nos ternary complex formation. Yeast expressing the Nos, Pum, or CP derivatives indicated were also transformed with plasmids that direct the synthesis of RNAs that contain either the MS2 hairpin (the CP-binding site), or the NRE, or both, as indicated in the drawing at left. The top plate monitors incorporation of Nos into a ternary complex, as shown schematically in Fig. 1A; the bottom plate monitors binding of Pum, as shown schematically in the drawing at right. (C) Northern blot analysis to assay the level of RNA accumulation in yeast. Samples of RNA from yeast transformed either with empty vector (−) or plasmids that direct synthesis of each of the indicated chimeric RNAs were probed with NRE sequences. Each lane contains approximately the same amount of low-molecular-weight RNA as determined by staining with ethidium bromide. Note that the NRE+ RNA (i.e., lacking the MS2 hairpin) accumulates to a higher level than do the NRE/MS2 chimeric RNAs. Compared with yeast expressing the NRE+/MS2 chimera (also cotransformed with Pum–DBD and Nos–AD plasmids), yeast expressing the NRE+ RNA grow on His− media in the presence of higher levels of the HIS3 competitor 3-aminotriazole and express higher levels of LacZ (not shown). These observations suggest that the concentration of RNA substrate is limiting in the yeast ternary complex assay.
Figure 2
Bases in the center of the NRE are required to recruit Nos. (A) Sequence of NRE+ and mutants that reduce its activity in embryos (Murata and Wharton 1995; Wharton et al. 1998). Bases conserved among NREs in bcd and hb genes from various Drosophila species are boxed. Mutations above reduce binding of Pum and those below prevent incorporation of Nos into a ternary complex. Mutant NREs used in these studies, each bearing a dinucleotide substitution, are named according to their position. (B) In vivo yeast assays of Pum/NRE/Nos ternary complex formation. Yeast expressing the Nos, Pum, or CP derivatives indicated were also transformed with plasmids that direct the synthesis of RNAs that contain either the MS2 hairpin (the CP-binding site), or the NRE, or both, as indicated in the drawing at left. The top plate monitors incorporation of Nos into a ternary complex, as shown schematically in Fig. 1A; the bottom plate monitors binding of Pum, as shown schematically in the drawing at right. (C) Northern blot analysis to assay the level of RNA accumulation in yeast. Samples of RNA from yeast transformed either with empty vector (−) or plasmids that direct synthesis of each of the indicated chimeric RNAs were probed with NRE sequences. Each lane contains approximately the same amount of low-molecular-weight RNA as determined by staining with ethidium bromide. Note that the NRE+ RNA (i.e., lacking the MS2 hairpin) accumulates to a higher level than do the NRE/MS2 chimeric RNAs. Compared with yeast expressing the NRE+/MS2 chimera (also cotransformed with Pum–DBD and Nos–AD plasmids), yeast expressing the NRE+ RNA grow on His− media in the presence of higher levels of the HIS3 competitor 3-aminotriazole and express higher levels of LacZ (not shown). These observations suggest that the concentration of RNA substrate is limiting in the yeast ternary complex assay.
Figure 3
The carboxy-terminal portion of Nos mediates recruitment into the ternary complex. (A) Drawings (to scale) of Nos derivatives tested for ternary complex formation in yeast. The conserved Cys/His domain is filled in. Results, shown in B, are summarized at right. Note that deletion of residues at the amino terminus of Nos (i.e., amino acids 1–42 at the junction with the AD) abolishes activity for unknown reasons, even though these are dispensable for nos function in vivo, as determined using a suitably modified transgene (data not shown). Note also that residues 1–42 are not present in the His6–Nos protein used to assay ternary complex formation in vitro (Fig. 6). (C) Western blot of yeast extracts prepared from transformants expressing each of the indicated Nos–AD derivatives. The blots were probed with a monoclonal anti-HA antibody that recognizes a vector-encoded epitope tag. Each derivative (arrowhead) accumulates to a level equal to or greater than the level of the wild-type (WT) fusion, with the exception of the ΔN3 derivative, which accumulates to a slightly lower level. For the blot at right, the antibody was preadsorbed and the background is lower as a result.
Figure 3
The carboxy-terminal portion of Nos mediates recruitment into the ternary complex. (A) Drawings (to scale) of Nos derivatives tested for ternary complex formation in yeast. The conserved Cys/His domain is filled in. Results, shown in B, are summarized at right. Note that deletion of residues at the amino terminus of Nos (i.e., amino acids 1–42 at the junction with the AD) abolishes activity for unknown reasons, even though these are dispensable for nos function in vivo, as determined using a suitably modified transgene (data not shown). Note also that residues 1–42 are not present in the His6–Nos protein used to assay ternary complex formation in vitro (Fig. 6). (C) Western blot of yeast extracts prepared from transformants expressing each of the indicated Nos–AD derivatives. The blots were probed with a monoclonal anti-HA antibody that recognizes a vector-encoded epitope tag. Each derivative (arrowhead) accumulates to a level equal to or greater than the level of the wild-type (WT) fusion, with the exception of the ΔN3 derivative, which accumulates to a slightly lower level. For the blot at right, the antibody was preadsorbed and the background is lower as a result.
Figure 3
The carboxy-terminal portion of Nos mediates recruitment into the ternary complex. (A) Drawings (to scale) of Nos derivatives tested for ternary complex formation in yeast. The conserved Cys/His domain is filled in. Results, shown in B, are summarized at right. Note that deletion of residues at the amino terminus of Nos (i.e., amino acids 1–42 at the junction with the AD) abolishes activity for unknown reasons, even though these are dispensable for nos function in vivo, as determined using a suitably modified transgene (data not shown). Note also that residues 1–42 are not present in the His6–Nos protein used to assay ternary complex formation in vitro (Fig. 6). (C) Western blot of yeast extracts prepared from transformants expressing each of the indicated Nos–AD derivatives. The blots were probed with a monoclonal anti-HA antibody that recognizes a vector-encoded epitope tag. Each derivative (arrowhead) accumulates to a level equal to or greater than the level of the wild-type (WT) fusion, with the exception of the ΔN3 derivative, which accumulates to a slightly lower level. For the blot at right, the antibody was preadsorbed and the background is lower as a result.
Figure 4
Human Pum binds to the NRE but does not recruit Nos. Shown are drawings (to scale) of the RNA-binding domains of Drosophila Pum (DmPum), human Pum (HsPum), and various chimeric proteins. The results of yeast in vivo interaction assays are summarized at right, with specific binding to NRE+ (and not NRE21) assayed as in Fig. 2B and recruitment of Nos into a ternary complex assayed as in Fig. 1A. Each of the eight repeats that comprise the core of the RNA-binding domain is indicated by a box, which is labeled only on the first line for clarity.
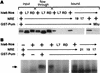
Figure 5
A Pum mutant that cannot recruit Nos does not regulate hb mRNA in vivo. (A) In vivo yeast assays of NRE binding (left) and Nos recruitment (right) by wild-type Pum and two mutants. Experimental designs for these experiments are shown schematically in Figs. 2B and 1A, respectively. Yeast express the chimeric molecules shown below as well as the Pum derivatives shown at left. (B) Western blot of samples prepared from yeast transformed with empty vector (−) or plasmids encoding each of the indicated Pum/DBD fusions probed with anti-Pum antibodies. Although there is less Pum680 than Pum+ in vivo, evidently the level of this mutant protein is saturating, as both transformants grow at a similar rate on His− plates containing various 3-AT concentrations, and both stimulate the lacZ reporter to a similar extent (data not shown). Note that overproduction of Pum+ also results in the accumulation of lower-molecular-weight (i.e., <49 kD) apparent breakdown products, which are visible for PumMlu in lane 3 (not shown). (C) Western blot of control wild-type (WT) or transgenic embryos expressing the RNA-binding domain fragments described in the text and indicated above. The blot was probed with anti-Pum antibodies, which recognize both the endogenous full-length protein (upper arrow) as well as the protein encoded by the transgene (lower arrow). (D) UV cross-linking of extracts prepared from wild-type control or transgenic embryos expressing either Pum+ or PumMlu RNA-binding domain derivatives, as indicated. In each pair of lanes, an aliquot of each extract was incubated with labeled NRE+ (left) or NRE21 (right) RNA. The upper arrow indicates the endogenous full-length Pum protein and the lower arrow indicates the transgene-encoded RNA-binding domain. (E) Each line shows the distribution of Hb in the early embryo in a midline, lateral view (left) and a dark-field photograph of the cuticle secreted by the mature embryo in the vitelline membrane in a ventral surface view (right). Embryos are derived from _pum_− females (top row) or _pum_− females that express the Pum+ or PumMlu RNA-binding domain fragments from transgenes (middle and bottom rows, as indicated). Embryos are oriented anterior at left. Note that only in the posterior half of the embryo is Hb accumulation derived solely from translation of maternal mRNA; in the anterior, Bicoid-dependent zygotic transcription of hb also contributes. Note also that in the pumET3/pumMsc background weak residual pum activity is evident at earlier stages of embryogenesis. In the right column, abdominal segmentation (prominent bands of thick hairs that appear as white dots) is apparent only in the middle panel.
Figure 6
Ternary complex formation in vitro. (A) Purified His6-Nos is visualized by Western blot using an antibody that recognizes the amino-terminal Xpress tag. This protein is degraded during purification from bacteria, yielding a doublet. In each reaction, either the wild-type molecule (+) or the indicated mutant derivative was incubated in a binding reaction, and ternary complexes were captured on glutathione–agarose beads, as described in Materials and Methods. Note that only the longer His6–Nos derivative is incorporated into ternary complexes. Lanes 1–3 contain 10% of the input, lanes 4–6 contain 10%–15% of the flowthrough, and lanes 7–14 contain 100% of the bound fraction. (B) The results of UV cross-linking experiments in which covalent RNA–protein adducts are detected by autoradiography following preincubation of the indicated mixture of molecules (above each lane) and UV irradiation. The RNA cross-links to both Pum and Nos; because the latter reaction is very inefficient, the signal from the Pum–RNA adduct was removed and only a portion of the autoradiogram is shown. The Nos–RNA adduct (arrow) is a doublet, presumably because of heterogeneity in the length of the RNA. Note that an impurity in the His6–Nos preparation cross-links nonspecifically to both wild-type and mutant RNAs (visible at bottom).
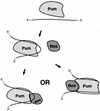
Figure 7
Models of ternary complex formation in the embryo. At left, Nos is recruited by a combination of weak but specific protein–protein and protein–RNA contacts. At right, Nos is recruited primarily by specific contacts to a surface of Pum that is competent only on binding to the NRE, although nonspecific contacts between Nos and the RNA may also stabilize the ternary complex.
Similar articles
- Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos.
Murata Y, Wharton RP. Murata Y, et al. Cell. 1995 Mar 10;80(5):747-56. doi: 10.1016/0092-8674(95)90353-4. Cell. 1995. PMID: 7889568 - The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence.
Zamore PD, Bartel DP, Lehmann R, Williamson JR. Zamore PD, et al. Biochemistry. 1999 Jan 12;38(2):596-604. doi: 10.1021/bi982264s. Biochemistry. 1999. PMID: 9888799 - The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins.
Zamore PD, Williamson JR, Lehmann R. Zamore PD, et al. RNA. 1997 Dec;3(12):1421-33. RNA. 1997. PMID: 9404893 Free PMC article. - Posttranscriptional regulation in Drosophila oocytes and early embryos.
Lasko P. Lasko P. Wiley Interdiscip Rev RNA. 2011 May-Jun;2(3):408-16. doi: 10.1002/wrna.70. Epub 2011 Jan 28. Wiley Interdiscip Rev RNA. 2011. PMID: 21957026 Review. - Translational repression: a duet of Nanos and Pumilio.
Parisi M, Lin H. Parisi M, et al. Curr Biol. 2000 Jan 27;10(2):R81-3. doi: 10.1016/s0960-9822(00)00283-9. Curr Biol. 2000. PMID: 10662662 Review.
Cited by
- Direct inhibition of Pumilo activity by Bam and Bgcn in Drosophila germ line stem cell differentiation.
Kim JY, Lee YC, Kim C. Kim JY, et al. J Biol Chem. 2010 Feb 12;285(7):4741-6. doi: 10.1074/jbc.M109.002014. Epub 2009 Dec 14. J Biol Chem. 2010. PMID: 20018853 Free PMC article. - Sex Determination in Nematode Germ Cells.
Ellis RE. Ellis RE. Sex Dev. 2022;16(5-6):305-322. doi: 10.1159/000520872. Epub 2022 Feb 16. Sex Dev. 2022. PMID: 35172320 Free PMC article. Review. - Pumilio Puf domain RNA-binding proteins in Arabidopsis.
Abbasi N, Park YI, Choi SB. Abbasi N, et al. Plant Signal Behav. 2011 Mar;6(3):364-8. doi: 10.4161/psb.6.3.14380. Epub 2011 Mar 1. Plant Signal Behav. 2011. PMID: 21350339 Free PMC article. Review. - Novel functions of the ubiquitin-independent proteasome system in regulating Xenopus germline development.
Hwang H, Jin Z, Krishnamurthy VV, Saha A, Klein PS, Garcia B, Mei W, King ML, Zhang K, Yang J. Hwang H, et al. Development. 2019 Apr 23;146(8):dev172700. doi: 10.1242/dev.172700. Development. 2019. PMID: 30910828 Free PMC article. - Hermes (Rbpms) is a Critical Component of RNP Complexes that Sequester Germline RNAs during Oogenesis.
Aguero T, Zhou Y, Kloc M, Chang P, Houliston E, King ML. Aguero T, et al. J Dev Biol. 2016 Mar;4(1):2. doi: 10.3390/jdb4010002. Epub 2016 Jan 19. J Dev Biol. 2016. PMID: 26998427 Free PMC article.
References
- Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes & Dev. 1992;6:2312–2326. - PubMed
- Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126:659–669. - PubMed
- Dahanukar A, Wharton RP. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes & Dev. 1996;10:2610–2620. - PubMed
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous