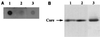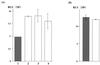Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation - PubMed (original) (raw)
Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation
T Shimoike et al. J Virol. 1999 Dec.
Abstract
To clarify the binding properties of hepatitis C virus (HCV) core protein and its viral RNA for the encapsidation, morphogenesis, and replication of HCV, the specific interaction of HCV core protein with its genomic RNA synthesized in vitro was examined in an in vivo system. The positive-sense RNA from the 5' end to nucleotide (nt) 2327, which covers the 5' untranslated region (5'UTR) and a part of the coding region of HCV structural proteins, interacted with HCV core protein, while no interaction was observed in the same region of negative-sense RNA and in other regions of viral and antiviral sense RNAs. The internal ribosome entry site (IRES) exists around the 5'UTR of HCV; therefore, the interaction of the core protein with this region of HCV RNA suggests that there is some effect on its cap-independent translation. Cells expressing HCV core protein were transfected with reporter RNAs consisting of nt 1 to 709 of HCV RNA (the 5'UTR of HCV and about two-thirds of the core protein coding regions) followed by a firefly luciferase gene (HCV07Luc RNA). The translation of HCV07Luc RNA was suppressed in cells expressing the core protein, whereas no significant suppression was observed in the case of a reporter RNA possessing the IRES of encephalomyocarditis virus followed by a firefly luciferase. This suppression by the core protein occurred in a dose-dependent manner. The expression of the E1 envelope protein of HCV or beta-galactosidase did not suppress the translation of both HCV and EMCV reporter RNAs. We then examined the regions that are important for suppression of translation by the core protein and found that the region from nt 1 to 344 was enough to exert this suppression. These results suggest that the HCV core protein interacts with viral genomic RNA at a specific region to form nucleocapsids and regulates the expression of HCV by interacting with the 5'UTR.
Figures
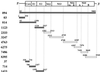
FIG. 1
HCV RNAs used in this study. The gene organization of HCV is shown at the top. The gray and white bars indicate the RNAs used in the experiments. Numbers on the bars indicate the positions of both ends of the RNAs. Gray bars indicate regions of viral sense RNA exhibiting a specific interaction with the core protein, as shown in Fig. 2 to 4.
FIG. 2
Specific interaction of HCV core protein with HCV RNA. HepG2 cells were infected with AcCA39 at an MOI of 50, transfected with +094 RNA (lane 1), −094 RNA (lane 2), or a transcript derived from pBlue (lane 3), and immunoprecipitated with anticore antibody. DIG-labeled RNAs were extracted from the immunoprecipitates and detected by dot blotting (A). HCV core protein was detected in the immunoprecipitates by Western blotting (B).
FIG. 3
Interaction of various regions of HCV RNA with core protein. HepG2 cells infected with AcCA39 were transfected with DIG-labeled 03, 014, 1123, 2333, 3247, 4763, 6275, 7486, or 8395 RNA. + and − indicate cells transfected with positive- and negative-sense RNAs, respectively. (A) Northern blots of RNAs extracted from the immunoprecipitates; (B) DIG-labeled RNAs recovered from the cells before immunoprecipitation.
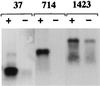
FIG. 4
HCV RNA regions responsible for a specific interaction with core protein. HepG2 cells infected with AcCA39 were transfected with DIG-labeled 37, 714, or 1423 RNA. + and − indicate cells transfected with positive- and negative-sense RNAs, respectively. RNAs extracted from the immunoprecipitates were analyzed by Northern blotting.
FIG. 5
Structures of reporter RNAs. HCVLuc consists of the 5′UTR of HCV followed by the luciferase gene. HCV07Luc is composed of the 5′UTR of HCV and a part of the coding region of the core protein (nt 1 to 709) followed by the luciferase gene. HCV09Luc consists of the 5′UTR of HCV, the whole core protein coding region, and part of the E1 protein coding region (nt 1 to 924) followed by the luciferase gene. EMCVLuc consist of the 5′UTR (nt 271 to 831) of EMCV followed by the luciferase gene.
FIG. 6
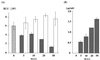
Suppression of translation of HCV RNA by core protein expression. (A) HepG2 cells infected with AcCA39 (lane 1, gray bar), AcCA816 (lane 2), AcCAlacZ (lane 3), or AcCAG (lane 4) at an MOI of 20 were transfected with HCV07Luc together with the internal standard, capped RLuc RNA. (B) HepG2 cells infected with AcCA39 (gray bar) or AcCAG (open bar) at an MOI of 20 were transfected with EMCVLuc together with the capped RLuc RNA. The activities of both firefly and RLuc were measured by a luminometer. Relative luciferase activity (RLU) is shown after normalization with that of the RLuc, which was used as an internal standard. Relative activities were determined in at least three independent experiments, each conducted with triplicate samples. Standard deviations are represented by vertical lines.
FIG. 7
Dose-dependent suppression of HCV RNA translation by core protein. (A) HepG2 cells infected with AcCA39 (gray bars) or AcCAG (open bars) at MOIs of 5 to 50 were cotransfected with HCV07Luc and capped RLuc RNAs. Cells were lysed at 6 h posttransfection, and both firefly and RLuc activities (RLU) were measured. The hatched bar indicates mock-infected cells. Relative activities (RLU) were determined as described for Fig. 6. (B) The amount of core protein in the same sample used for the luciferase assay was measured by ELISA.
FIG. 8
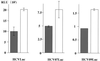
HCV core protein suppresses the translation of RNA possessing the 5′UTR of HCV. HepG2 cells infected with AcCA39 (gray bars) or AcCAG (open bars) at an MOI of 20 were cotransfected with HCVLuc, HCV07Luc, or HCV09Luc RNA together with the internal standard, capped RLuc RNA. Relative activities (RLU) were determined as described for Fig. 6.
Similar articles
- A Novel Cis-Acting RNA Structural Element Embedded in the Core Coding Region of the Hepatitis C Virus Genome Directs Internal Translation Initiation of the Overlapping Core+1 ORF.
Vassilaki N, Frakolaki E, Kalliampakou KI, Sakellariou P, Kotta-Loizou I, Bartenschlager R, Mavromara P. Vassilaki N, et al. Int J Mol Sci. 2020 Sep 22;21(18):6974. doi: 10.3390/ijms21186974. Int J Mol Sci. 2020. PMID: 32972019 Free PMC article. - Hepatitis C virus core protein acts as a trans-modulating factor on internal translation initiation of the viral RNA.
Boni S, Lavergne JP, Boulant S, Cahour A. Boni S, et al. J Biol Chem. 2005 May 6;280(18):17737-48. doi: 10.1074/jbc.M501826200. Epub 2005 Mar 9. J Biol Chem. 2005. PMID: 15760888 - [Structure and function of the non-coding regions of hepatitis C viral RNA].
Dutkiewicz M, Swiqtkowska A, Ciesiołka J. Dutkiewicz M, et al. Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish. - RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis.
Piñeiro D, Martinez-Salas E. Piñeiro D, et al. Viruses. 2012 Oct 19;4(10):2233-50. doi: 10.3390/v4102233. Viruses. 2012. PMID: 23202462 Free PMC article. Review.
Cited by
- HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions.
Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K. Hafrén A, et al. Plant Cell. 2010 Feb;22(2):523-35. doi: 10.1105/tpc.109.072413. Epub 2010 Feb 12. Plant Cell. 2010. PMID: 20154150 Free PMC article. - Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles.
Clayton RF, Owsianka A, Aitken J, Graham S, Bhella D, Patel AH. Clayton RF, et al. J Virol. 2002 Aug;76(15):7672-82. doi: 10.1128/jvi.76.15.7672-7682.2002. J Virol. 2002. PMID: 12097581 Free PMC article. - Apo B100 similarities to viral proteins suggest basis for LDL-DNA binding and transfection capacity.
Guevara J Jr, Prashad N, Ermolinsky B, Gaubatz JW, Kang D, Schwarzbach AE, Loose DS, Guevara NV. Guevara J Jr, et al. J Lipid Res. 2010 Jul;51(7):1704-18. doi: 10.1194/jlr.M003277. Epub 2010 Feb 19. J Lipid Res. 2010. PMID: 20173184 Free PMC article. - Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta.
Zhang XP, Liu DS, Yan T, Fang XD, Dong K, Xu J, Wang Y, Yu JL, Wang XB. Zhang XP, et al. PLoS Pathog. 2017 Jul 20;13(7):e1006522. doi: 10.1371/journal.ppat.1006522. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28727810 Free PMC article.
References
- Aizaki H, Aoki Y, Harada T, Ishii K, Suzuki T, Nagamori S, Toda G, Matsuura Y, Miyamura T. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology. 1998;27:621–627. - PubMed
- Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, Meeks E L, Beach M J. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. - PubMed
- Aoki Y, Aizaki H, Shimoike T, Tani H, Ishii K, Saito I, Matsuura Y, Miyamura T. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology. 1998;250:140–150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous