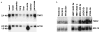TIN2, a new regulator of telomere length in human cells - PubMed (original) (raw)
TIN2, a new regulator of telomere length in human cells
S H Kim et al. Nat Genet. 1999 Dec.
Abstract
Telomeres are DNA-protein structures that cap linear chromosomes and are essential for maintaining genomic stability and cell phenotype. We identified a novel human telomere-associated protein, TIN2, by interaction cloning using the telomeric DNA-binding-protein TRF1 as a bait. TIN2 interacted with TRF1 in vitro and in cells, and co-localized with TRF1 in nuclei and metaphase chromosomes. A mutant TIN2 that lacks amino-terminal sequences effects elongated human telomeres in a telomerase-dependent manner. Our findings suggest that TRF1 is insufficient for control of telomere length in human cells, and that TIN2 is an essential mediator of TRF1 function.
Figures
Fig. 1
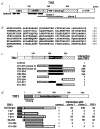
Sequence and structural characteristics of human TIN2. a, Structural features of TIN2. Shown are regions corresponding to the cDNA inserts recovered from the two-hybrid screen (clone 1, aa 147–275; clone 2, aa 196–354), the basic and acidic regions, potential helical structures and TRF1-binding domain. b, Deduced amino acid sequence of TIN2. c, TIN2 domains that interact with TRF1. We transformed TINF2 cDNA fragments (encoding the indicated amino acids) in pGAD-424 into yeast with pGBT9 containing TERF1 cDNA, and assessed interaction by a luminescent β-galactosidase assay. Control luminescence (interaction of pGAD-424 with pGBT9-TRF1) was 0.1–0.2 β-galactosidase U, and given a value of 1. We analysed 3–5 transformants for each determination. c, TRF1 domains that interact with TIN2. Depicted is TRF1, showing the tankyrase-binding and homodimerization domains. We transformed TERF1 cDNA fragments (encoding the indicated amino acids) in pGBT9 into yeast with pGAD10 containing no insert (control), TINF2 clone 1, TINF2 clone 2 or full-length TERF1 cDNA, and assessed interaction by luminescent β-galactosidase assay. Control luminescence (interaction with insertless pGAD10) was 0.1–0.2 β-galactosidase U, and given a value of 1. We analysed 3–5 transformants for each determination.
Fig. 2
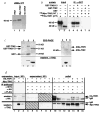
TIN2 interacts with TRF1 in vitro and in cells. a, Translation products of TINF2 and Myc_–_TINF2 cDNAs. We transcribed and translated with 35S-methionine the TINF2 and Myc_–_TINF2 cDNAs in vitro, and analysed the translation products by SDS–PAGE. Lane 1, TINF2 cDNA; lane 2, Myc_–_TINF2 cDNA (lacking the 5′ UTR); lane 3, no cDNA control. b, TIN2 binds TRF1, but not TIN2, in vitro. We generated radiola-belled Myc–TIN2 and HA–TRF1 proteins by in vitro translation, and analysed 2 μl of the reactions by SDS–PAGE (lanes 1,2). In parallel, we incubated 5 μl of the reactions with 20 ng of GST (lanes 3,6), GST–TIN2 (lanes 4,7) or GST–TIN2-13 (lanes 5,8), immunoprecipitated the GST complexes, eluted proteins in the immune complexes into SDS–PAGE sample buffer and analysed 50% of the eluate by SDS–PAGE. c, Recombinant proteins and anti-TIN2 antibody. We expressed GST (lane 1), GST–TIN2 (lane 2), and GST–TIN2-13 (lane 3) in Escherichia coli, and 6His–TRF1 (lane 4), 6His–TIN2 (lane 5) and 6His–control (6His plus 36-bp vector sequence; lane 6) proteins using baculovirus and insect cells, purified them from cell lysates by glutathione (GST proteins) or nickel (6His proteins) chromatography, and analysed them by SDS–PAGE (top) and western blot (bottom) using affinity-purified anti-TIN2. d, TIN2 and TRF1 interact in cells. We prepared pre-cleared lysates from HT1080 cells that overexpress HA–TRF1, Myc–TIN2 or both (expression), subjected 20% of each lysate to SDS–PAGE (lanes 1-3), immunoprecipitated the remaining 80% with mouse monoclonal anti-HA, anti-Myc or anti-FLAG (control) antibodies (IP antibody), and collected immune complexes on protein A–Sepharose beads. We analysed 20% of each depleted supernatant by SDS–PAGE (lanes 4–9). We released proteins in the immune complexes into SDS sample buffer, subjected them to SDS–PAGE (lanes 10–16), and analysed them by western blot using affinity-purified rabbit polyclonal anti-HA (top; western, anti-HA) or anti-TIN2 (bottom; western, anti-TIN2) antibodies. Indicated are the positions of HA–TRF1, Myc–TIN2 and cross-reacting IgG heavy chains.
Fig. 3
Truncated TIN2 proteins extend telomere length. a, TIN2 proteins used in this experiment. Shown are the N-terminal regions N1 (aa 1–120) and N2 (aa 120–196), TRF1-interaction domain (TRF-int) and C-terminal domain (C1). b, TIN2 expression. We infected HT1080 cells with control or the indicated Myc–TIN2-expressing retroviruses, and analysed cell lysates prepared 3–6 PD (lanes 3,5,7,9) and 60 PD (lanes 4,6,8,10) after selection by western blot, using anti-TIN2 or anti-tubulin (control) antibodies. c,d, Effects on TRF length. We infected HT1080 cells with control or the indicated Myc–TIN2-expressing retroviruses, selected for virus-expressing cells and permitted the cells to proliferate for the PD number indicated above each lane before DNA was isolated and analysed for TRF length. c, Hybridization from one experiment. d, Average intensity of the peak hybridization signal versus PD number from two or three independent experiments.
Fig. 4
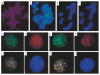
TIN2 subcellular localization. We fixed HT1080 cells, uninfected or expressing HA–TRF1, Myc–TIN2 or Myc–TIN2-13 retroviruses, while proliferating, or after treatment with colcemid to obtain metaphase chromosomes, stained them with anti-HA, anti-Myc or anti-TIN2 antibodies and applied secondary antibodies (FITC (green fluorescence)- or Texas Red (red fluorescence)-conjugated anti-rabbit or anti-mouse IgG). We visualized DNA by DAPI staining (blue fluorescence), and photographed the cells and chromosomes using a digital camera, merging the images where indicated. a, Metaphase chromosomes from uninfected cell stained with anti-TIN2 (endogenous TIN2) antibody. b, Meta-phase chromosomes from Myc–TIN2-expressing cell stained with anti-Myc (retroviral TIN2) antibody. c, Interphase nucleus of an HA–TRF-1/Myc–TIN2-expressing cell stained with anti-Myc (retroviral TIN2) antibody. d, Inter-phase nucleus of the same HA–TRF-1/Myc–TIN2-expressing cell stained with anti-HA (retroviral TRF1) antibody. e, Co-localization of HA–TRF1 and Myc–TIN2 in nucleus shown in (c) and (d) (merged image). f, DAPI staining of nucleus shown in (c_–_e). g, Inter-phase nucleus of another HA–TRF-1/Myc–TIN2-expressing cell stained with anti-Myc (retroviral TIN2) antibody. h, Interphase nucleus of the same HA–TRF-1/Myc–TIN2-expressing cells stained with anti-HA (retroviral TRF1) antibody. i, Co-localization of HA–TRF1/Myc–TIN2 in nucleus shown in (g) and (h) (merged image). j, DAPI staining of nucleus shown in (g_–_i). k, Metaphase chromosomes from HA–TRF1/Myc–TIN2-13–expressing cell stained with anti-HA antibody (telomeric localization of TRF1 in the presence of TIN2-13). l, Metaphase chromosomes from HA–TRF1/Myc–TIN2-13–expressing cells stained with anti-Myc antibody (telomeric localization of TIN2-13).
Fig. 5
Expression pattern of TINF2 mRNA. a, Expression in human tissues. We analysed RNA from the indicated human tissues by northern blot to detect the TINF2 and ACTB (β-actin) mRNAs. Indicated are the 2.4-kb TINF2, 2.0-kb ACTB and 1.8-kb cross-hybridizing cardiac and skeletal muscle actin mRNAs. b, Expression in human cells. We analysed RNA from the indicated cell cultures by northern blot to detect the TINF2 and RPL10 (control) mRNAs.
Fig. 6
Telomerase dependence. a, Telomerase activity. We infected WI-38 (lanes 1,2) or hTERT (hT)-expressing WI-38 (lanes 3–8) cells with control (Lx, lanes 1–4), TIN2 (lanes 5,6) or TIN2-13 (lanes 7,8) retroviruses, selected for virus-expressing cells and prepared cell lysates. We analysed cell lysate volumes equivalent to equal numbers of cells for telomerase activity by TRAP assay. neg, extracts heated to 85 °C before assay. b, TRF length. We infected WI-38 cells at PD 29 (lane 1) with pBabe control (lanes 2,4,6,7) or hTERT-expressing (lanes 3,5,8,9) virus. After five to six PD, we superinfected the cells with viruses expressing LXSN control (Lx; lanes 2,3), TIN2 (lanes 4,5) or TIN2-13 (lanes 6–9). We isolated DNA at the indicated PD levels, and analysed the DNA for TRF length. c, TIN2 does not inhibit telomerase activity in vitro. We prepared extracts from HT1080 cells (lane 2) and mixed equal aliquots with 20 ng GST (lane 4), or 1 (lane 5), 5 (lane 6) or 20 (lane 7) ng GST–TIN2. We incubated the extracts for 10 min at 4 °C before assaying for telomerase activity. pos, positive extract from the assay kit; neg, extract heated to 85 °C.
Fig. 7
TIN2-13 does not displace TRF1. a, 6His–TRF1 and 6His–TIN2 DNA-binding activity. We incubated recombinant proteins with a double-stranded TTAGGG6 probe, and analysed protein-DNA complexes by EMSA. Lane 1, 6-His–TRF1 (150 ng) analysed alone; lane 2, 6-His–TRF1 (150 ng) plus 100-fold excess unlabelled mutant [TTAGGC]7 probe (Mut competitor); lane 3, 6-His–TRF1 (150 ng) plus 100-fold excess unlabelled wild-type [TTAGGG]7 probe (WT competitor; lane 3); lane 4, 150 ng 6His–TRF1 plus an equal volume of 6His–control protein (6His plus 36 bp vector sequence, expressed and purified identically to 6His–TRF1 and 6His–TIN2); lanes 5–8, 150 ng 6His–TRF1 plus 10, 40, 150 or 0 ng 6His–TIN2; lane 9, 150 ng 6His–TIN2 alone. The TRF1-specific band is indicated. b, GST–TIN2 and GST–TIN2-13 binding activity, and interaction with TRF1. We incubated recombinant proteins, without or with nuclear extract (NE) from HA–TRF1-expressing HT1080 cells or antibodies, with a double-stranded TTAGGG13 probe, and analysed protein-DNA complexes by EMSA. Lane 1, 20 ng GST–TIN2 alone; lane 2, 20 ng GST–TIN2-13 alone; lane 3, NE alone; lane 4, NE plus 100-fold excess unlabelled [TTAGGG]7 (WT competitor); lane 5, NE plus 0.2 μg anti-HA antibody; lane 6, NE plus 0.2 μg anti-Myc (control) antibody; lanes 7–11, NE plus 20 ng GST, GST–TIN2-13, GST–TIN2-WT or TIN2 aa 1–196 (lacking the TRF1-binding domain) fused to GST (GST–TIN2-Nter); lane 12, NE; lane 13, NE plus 20 ng GST–TIN2-13; lane 14, NE plus 20 ng GST–TIN2-13 plus 0.2 μg anti-GST; lane 15, NE plus 20 ng GST–TIN2-13 plus 0.2 μg anti-Myc (control); lane 16, NE plus 20 ng GST–TIN2-13 plus 0.2 μg anti-HA. The TRF1-specific band, and a non-specific band (ns) present in some of the gels, is indicated.
Comment in
- At the end of the millennium, a view of the end.
Shay JW. Shay JW. Nat Genet. 1999 Dec;23(4):382-3. doi: 10.1038/70480. Nat Genet. 1999. PMID: 10581016 No abstract available.
Similar articles
- Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas.
Matsutani N, Yokozaki H, Tahara E, Tahara H, Kuniyasu H, Haruma K, Chayama K, Yasui W, Tahara E. Matsutani N, et al. Int J Oncol. 2001 Sep;19(3):507-12. Int J Oncol. 2001. PMID: 11494028 - TIN2 mediates functions of TRF2 at human telomeres.
Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. Kim SH, et al. J Biol Chem. 2004 Oct 15;279(42):43799-804. doi: 10.1074/jbc.M408650200. Epub 2004 Aug 3. J Biol Chem. 2004. PMID: 15292264 - TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex.
Ye JZ, de Lange T. Ye JZ, et al. Nat Genet. 2004 Jun;36(6):618-23. doi: 10.1038/ng1360. Epub 2004 May 9. Nat Genet. 2004. PMID: 15133513 - Role of Pin2/TRF1 in telomere maintenance and cell cycle control.
Zhou XZ, Perrem K, Lu KP. Zhou XZ, et al. J Cell Biochem. 2003 May 1;89(1):19-37. doi: 10.1002/jcb.10496. J Cell Biochem. 2003. PMID: 12682905 Review. - Regulation of telomerase by telomeric proteins.
Smogorzewska A, de Lange T. Smogorzewska A, et al. Annu Rev Biochem. 2004;73:177-208. doi: 10.1146/annurev.biochem.73.071403.160049. Annu Rev Biochem. 2004. PMID: 15189140 Review.
Cited by
- Telomere maintenance and the DNA damage response: a paradoxical alliance.
Harman A, Bryan TM. Harman A, et al. Front Cell Dev Biol. 2024 Oct 17;12:1472906. doi: 10.3389/fcell.2024.1472906. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39483338 Free PMC article. - TRF2-RAP1 represses RAD51-dependent homology-directed telomere repair by promoting BLM-mediated D-loop unwinding and inhibiting BLM-DNA2-dependent 5'-end resection.
Liang F, Rai R, Sodeinde T, Chang S. Liang F, et al. Nucleic Acids Res. 2024 Sep 9;52(16):9695-9709. doi: 10.1093/nar/gkae642. Nucleic Acids Res. 2024. PMID: 39082275 Free PMC article. - TRF1 and TRF2: pioneering targets in telomere-based cancer therapy.
Kallingal A, Krzemieniecki R, Maciejewska N, Brankiewicz-Kopcińska W, Baginski M. Kallingal A, et al. J Cancer Res Clin Oncol. 2024 Jul 16;150(7):353. doi: 10.1007/s00432-024-05867-3. J Cancer Res Clin Oncol. 2024. PMID: 39012375 Free PMC article. Review. - Digital telomere measurement by long-read sequencing distinguishes healthy aging from disease.
Sanchez SE, Gu Y, Wang Y, Golla A, Martin A, Shomali W, Hockemeyer D, Savage SA, Artandi SE. Sanchez SE, et al. Nat Commun. 2024 Jun 18;15(1):5148. doi: 10.1038/s41467-024-49007-4. Nat Commun. 2024. PMID: 38890274 Free PMC article. - Structural Motifs at the Telomeres and Their Role in Regulatory Pathways.
Alanazi AFR, Parkinson GN, Haider S. Alanazi AFR, et al. Biochemistry. 2024 Apr 2;63(7):827-842. doi: 10.1021/acs.biochem.4c00023. Epub 2024 Mar 14. Biochemistry. 2024. PMID: 38481135 Free PMC article. Review.
References
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1995;66:1279–1287. - PubMed
- Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, function in silencing, cell cycle progression and chromosome stability. Genes Dev. 1995;9:2888–2902. - PubMed
- Marchand S, Buck SW, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by RAP1 protein. Genes Dev. 1996;10:1297–1309. - PubMed
- Campisi J. The biology of replicative senescence. Eur J Cancer. 1997;33:703–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AG000266/AG/NIA NIH HHS/United States
- AG09909/AG/NIA NIH HHS/United States
- R37 AG009909/AG/NIA NIH HHS/United States
- R56 AG009909/AG/NIA NIH HHS/United States
- AG00266/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous