Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage - PubMed (original) (raw)
Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage
R Khosravi et al. Proc Natl Acad Sci U S A. 1999.
Abstract
The p53 tumor-suppressor protein, a key regulator of cellular responses to genotoxic stress, is stabilized and activated after DNA damage. This process is associated with posttranslational modifications of p53, some of which are mediated by the ATM protein kinase. However, these modifications alone may not account in full for p53 stabilization. p53's stability and activity are negatively regulated by the oncoprotein MDM2, whose gene is activated by p53. Conceivably, p53 function may be modulated by modifications of MDM2 as well. We show here that after treatment of cells with ionizing radiation or a radiomimetic chemical, but not UV radiation, MDM2 is phosphorylated rapidly in an ATM-dependent manner. This phosphorylation is independent of p53 and the DNA-dependent protein kinase. Furthermore, MDM2 is directly phosphorylated by ATM in vitro. These findings suggest that in response to DNA strand breaks, ATM may promote p53 activity and stability by mediating simultaneous phosphorylation of both partners of the p53-MDM2 autoregulatory feedback loop.
Figures
Figure 1
Radiation-induced accumulation of p53(S15A). A plasmid encoding a mutant p53 protein, in which Ser15 was replaced by Ala (8), was transfected into ap29 murine p53-null cells (37) or into ap24 double-mutant cells lacking both p53 and Atm (37). After 48 hr, some of the cultures were treated with 7.5 Gy of gamma radiation and harvested 75 min later. Total cellular extracts were subjected to SDS/PAGE and immunoblot analysis by using a mixture of the anti-p53 antibodies PAb1801 and DO-1. The faster-migrating band observed in lane 3 is a truncated form of p53 often observed when p53 is produced at elevated levels.
Figure 2
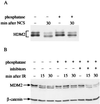
DNA damage-induced alterations of cellular HDM2. (A) HDM2 immunoprecipitated from cellular extracts of a human lymphoblastoid cell line (C3ABR) at various time points after addition of 80 ng/ml NCS. Immune complexes obtained by using the anti-HDM2 mAb IF2 were subjected to SDS/PAGE followed by immunoblotting with a mixture of mAbs 2A10, 1D6, 3G5, SMP14, and IF5. Note the rapid change toward accelerated electrophoretic mobility of HDM2 already apparent 5 min after treatment and the slower increase in its overall level that accompanies p53 accumulation (see Fig. 1_B_). (B) p53 levels in total cellular extracts of NCS-treated C3ABR lymphoblasts, detected by using the anti-p53 antibody DO-1. (C) HDM2 in total cellular extracts of p53-null Saos-2 cells. The blot was reacted with the same mixture of antibodies as in A. Note the shift of the major band toward a faster-migrating position. (D) Altered immunoreactivity of HDM2 after exposure to IR. Saos-2 cells were harvested at the indicated time points after treatment with 5 Gy of gamma radiation. Before their irradiation the cells were pretreated with 30 μM of the proteasome inhibitor MG132 for 2 hr to allow HDM2 accumulation. Total cellular extracts were immunoblotted with the two indicated anti-HDM2 mAbs. Note the differential decrease in immunoreactivity with 2A10 and the shift of the major (faster) band toward accelerated mobility observed by using 3F3. (E) Immunoreactivity of HDM2 with the 2A10 antibody decreases after exposure to ionizing but not UV radiation. Saos-2 cells were treated with either 10 Gy of gamma radiation or 30 J/m2 of UV radiation, and HDM2 was visualized as in Fig. 1_D_. In B_–_E, equal amounts of protein were loaded in all lanes in each. This is demonstrated in E by using a β-catenin antibody. In A, immunoprecipitates in each lane represent equal numbers of cells.
Figure 3
Alkaline phosphatase (AP) reverses damage-induced HDM2 or MDM2 modifications. (A) Immunoblot analysis of HDM2 immunoprecipitated from a human lymphoblastoid cell line (NL553) treated with 80 ng/ml NCS. NCS-treated cells were lysed 30 min after addition of the drug to the cultures. Cellular extracts were immunoprecipitated with the IF2 antibody, and the immune complexes were treated with shrimp alkaline phosphatase as described in Materials and Methods and subjected to SDS/PAGE and immunoblotting with the same antibody mixture as in Fig. 2_A_. Note the reversion of HDM2's mobility shift by AP treatment. (B) Immunoblot analysis of murine MDM2 using the 2A10 antibody. p53-null mouse fibroblasts (ap29) infected with a recombinant retrovirus encoding mouse MDM2 were treated with 7.5 Gy of ionizing radiation and harvested at various time points after irradiation. Cellular extracts were incubated for 30 min at 30°C, without (lanes 1–3) or with (lanes 4–6) calf intestine alkaline phosphatase (CIAP), or with CIAP and phosphatase inhibitors (50 mM NaF, 10 mM NaVO4; lanes 7–9). The band migrating slightly ahead of MDM2 is a nonspecific-background band also observed in MDM2-null cells.
Figure 4
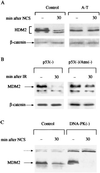
Damage-induced modifications of HDM2 or MDM2 are dependent on ATM but not DNA-PK. (A) Immunoblot analysis of HDM2 in total cellular extracts of control (L-39) and A-T (L-6) lymphoblasts before and after NCS treatment. The blots were reacted with the same antibody mixture as in Fig. 2_A_. (B) 2A10 immunoreactivity of MDM2 in total cellular extracts of murine ap29 cells lacking p53 and ap24 cells lacking both p53 and Atm (37) before and after treatment with 7.5 Gy of ionizing radiation. (C) 2A10 immunoreactivity of MDM2 in total cellular extracts of A9 murine control cells and 494 cells derived from the “slip” mice lacking DNA-PK activity (25) after NCS treatment. Note the complete loss of 2A10 immunoreactivity in DNA-PK-deficient cells. Cross-reacting bands (broken arrow) indicate relative protein amounts in different lanes.
Figure 5
In vitro phosphorylation of recombinant proteins by ATM. ATM was immunoprecipitated from control (L-40) and A-T (L-6) lymphoblastoid lines, and an immunoprecipitation-kinase reaction was performed for 15 min as described before (12). (A) Diagram showing various HDM2 fragments used as substrates (see Materials and Methods). The numbers in brackets denote the residues contained in each fragment. HDM2(1–115) was fused to an S tag, and all other fragments were produced as glutathione _S_-transferase (GST)-fusions. (B) In vitro phosphorylation of HDM2 and MDM2 fragments by ATM. Immunoprecipitated ATM was visualized by using immunoblotting. p53-N47, a polypeptide containing residues 1–47 of human p53 fused to the POU domain of the human transcription factor Oct-1 serving as a positive control substrate (12). Nonfused GST served as a negative control. (C) Reduction in immunoreactivity of recombinant HMD2 with the 2A10 mAb after in vitro phosphorylation by ATM. Full-length HDM2 was phosphorylated for 15 min by ATM and subsequently detected by immunoblotting with the antibodies 2A10 and 3F3.
Similar articles
- ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage.
Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M. Maya R, et al. Genes Dev. 2001 May 1;15(9):1067-77. doi: 10.1101/gad.886901. Genes Dev. 2001. PMID: 11331603 Free PMC article. - Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage.
Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, Shiloh Y. Pereg Y, et al. Proc Natl Acad Sci U S A. 2005 Apr 5;102(14):5056-61. doi: 10.1073/pnas.0408595102. Epub 2005 Mar 23. Proc Natl Acad Sci U S A. 2005. PMID: 15788536 Free PMC article. - ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage.
Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. Chen L, et al. EMBO J. 2005 Oct 5;24(19):3411-22. doi: 10.1038/sj.emboj.7600812. Epub 2005 Sep 15. EMBO J. 2005. PMID: 16163388 Free PMC article. - How to activate p53.
Caspari T. Caspari T. Curr Biol. 2000 Apr 20;10(8):R315-7. doi: 10.1016/s0960-9822(00)00439-5. Curr Biol. 2000. PMID: 10801407 Review. - Regulation of p53 in response to DNA damage.
Lakin ND, Jackson SP. Lakin ND, et al. Oncogene. 1999 Dec 13;18(53):7644-55. doi: 10.1038/sj.onc.1203015. Oncogene. 1999. PMID: 10618704 Review.
Cited by
- A Switch in p53 Dynamics Marks Cells That Escape from DSB-Induced Cell Cycle Arrest.
Tsabar M, Mock CS, Venkatachalam V, Reyes J, Karhohs KW, Oliver TG, Regev A, Jambhekar A, Lahav G. Tsabar M, et al. Cell Rep. 2020 Aug 4;32(5):107995. doi: 10.1016/j.celrep.2020.107995. Cell Rep. 2020. PMID: 32755587 Free PMC article. - Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress.
Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Tibbetts RS, et al. Genes Dev. 2000 Dec 1;14(23):2989-3002. doi: 10.1101/gad.851000. Genes Dev. 2000. PMID: 11114888 Free PMC article. - Mutational processes shape the landscape of TP53 mutations in human cancer.
Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, Howard TP, Takeda DY, Ly SH, Kim E, Gannon HS, Hurhula B, Sharpe T, Goodale A, Fritchman B, Steelman S, Vazquez F, Tsherniak A, Aguirre AJ, Doench JG, Piccioni F, Roberts CWM, Meyerson M, Getz G, Johannessen CM, Root DE, Hahn WC. Giacomelli AO, et al. Nat Genet. 2018 Oct;50(10):1381-1387. doi: 10.1038/s41588-018-0204-y. Epub 2018 Sep 17. Nat Genet. 2018. PMID: 30224644 Free PMC article. - The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation.
Zilfou JT, Hoffman WH, Sank M, George DL, Murphy M. Zilfou JT, et al. Mol Cell Biol. 2001 Jun;21(12):3974-85. doi: 10.1128/MCB.21.12.3974-3985.2001. Mol Cell Biol. 2001. PMID: 11359905 Free PMC article. - Twilight effects of low doses of ionizing radiation on cellular systems: a bird's eye view on current concepts and research.
Postiglione I, Chiaviello A, Palumbo G. Postiglione I, et al. Med Oncol. 2010 Jun;27(2):495-509. doi: 10.1007/s12032-009-9241-9. Epub 2009 Jun 6. Med Oncol. 2010. PMID: 19504191 Review.
References
- Levine A J. Cell. 1997;88:323–331. - PubMed
- Gottlieb T M, Oren M. Biochem Biophys Acta Rev Cancer. 1996;1287:77–102. - PubMed
- Agarwal M L, Taylor W R, Chemov M V, Chemova O B, Stark G R. J Biol Chem. 1998;273:1–4. - PubMed
- Giaccia A J, Kastan M B. Genes Dev. 1998;12:2973–2983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous