Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region - PubMed (original) (raw)
Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region
K Mahr et al. Appl Environ Microbiol. 2000 Jan.
Abstract
The catabolite control protein CcpA is a central regulator in low-G+C-content gram-positive bacteria. It confers carbon catabolite repression to numerous genes required for carbon utilization. It also operates as a transcriptional activator of genes involved in diverse phenomena, such as glycolysis and ammonium fixation. We have cloned the ccpA region of Lactobacillus pentosus. ccpA encodes a protein of 336 amino acids exhibiting similarity to CcpA proteins of other bacteria and to proteins of the LacI/GalR family of transcriptional regulators. Upstream of ccpA was found an open reading frame with similarity to the pepQ gene, encoding a prolidase. Primer extension experiments revealed two start sites of transcription for ccpA. In wild-type cells grown on glucose, mRNA synthesis occurred only from the promoter proximal to ccpA. In a ccpA mutant strain, both promoters were used, with increased transcription from the distant promoter, which overlaps a presumptive CcpA binding site called cre (for catabolite responsive element). This suggests that expression of ccpA is autoregulated. Determination of the expression levels of CcpA in cells grown on repressing and nonrepressing carbon sources revealed that the amounts of CcpA produced did not change significantly, leading to the conclusion that the arrangement of two promoters may ensure constant expression of ccpA under various environmental conditions. A comparison of the genetic structures of ccpA regions revealed that lactic acid bacteria possess the gene order pepQ-ccpA-variable while the genetic structure in bacilli and Staphylococcus xylosus is aroA-ccpA-variable-acuC.
Figures
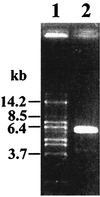
FIG. 1
Amplification product of 5.6 kb of the ccpA region of L. pentosus on a 1% agarose gel. Sizes of DNA fragments are indicated in kilobase pairs. Lane 1, DNA marker fragments; lane 2, 3 μl of a 50-μl-volume inverse PCR using oligonucleotides LPE1 and LPE2 and religated L. pentosus MD363 chromosomal DNA.
FIG. 2
Protein sequence of L. pentosus CcpA and comparison to a consensus sequence. Lowercase letters of the consensus sequence designate conserved amino acid residues specific for proteins of the LacI/GalR family of regulators, whereas uppercase letters designate amino acid residues specific for CcpA proteins (18).
FIG. 3
Genetic organization and transcriptional regulation of L. pentosus ccpA. (A) Genetic organization of the ccpA region. The size and orientation of the ORFs were deduced from the nucleotide sequence. The ccpA promoter region is depicted at the DNA sequence level. ccpA transcriptional start sites are in boldface and are marked by asterisks. Potential RBSs and cre motifs are underlined. Putative RNA polymerase binding sites (−10 region and −35 region) in the sequence are in boldface letters and underlined and are indicated with P1 and P2. The N-terminal protein sequences of pepQ and ccpA are shown. (B) Primer extension analysis of ccpA gene transcription of L. pentosus wild-type and ccpA mutant strains. Total RNA was prepared from cells grown on M medium supplemented with 50 mM glucose. Reverse transcription was carried out with end-labelled oligonucleotide LPE27. DNA sequencing reactions were performed with the same oligonucleotide and with pWH156 as template DNA. Primer extension products were analyzed on 6% polyacrylamide–urea gels. Lane 1, RNA (20 μg) from L. pentosus LPE4 (ccpA mutant); lane 2, RNA (20 μg) from L. pentosus MD363 (wild type). The sequence interpretations around the +1 sites (asterisks and arrows) of the two ccpA promoters are shown.
FIG. 3
Genetic organization and transcriptional regulation of L. pentosus ccpA. (A) Genetic organization of the ccpA region. The size and orientation of the ORFs were deduced from the nucleotide sequence. The ccpA promoter region is depicted at the DNA sequence level. ccpA transcriptional start sites are in boldface and are marked by asterisks. Potential RBSs and cre motifs are underlined. Putative RNA polymerase binding sites (−10 region and −35 region) in the sequence are in boldface letters and underlined and are indicated with P1 and P2. The N-terminal protein sequences of pepQ and ccpA are shown. (B) Primer extension analysis of ccpA gene transcription of L. pentosus wild-type and ccpA mutant strains. Total RNA was prepared from cells grown on M medium supplemented with 50 mM glucose. Reverse transcription was carried out with end-labelled oligonucleotide LPE27. DNA sequencing reactions were performed with the same oligonucleotide and with pWH156 as template DNA. Primer extension products were analyzed on 6% polyacrylamide–urea gels. Lane 1, RNA (20 μg) from L. pentosus LPE4 (ccpA mutant); lane 2, RNA (20 μg) from L. pentosus MD363 (wild type). The sequence interpretations around the +1 sites (asterisks and arrows) of the two ccpA promoters are shown.
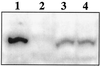
FIG. 4
Western blot analysis of L. pentosus grown on various carbon sources. A Western blot of a sodium dodecyl sulfate–7.5% polyacrylamide gel is shown after incubation with polyclonal antibodies derived against CcpA of B. megaterium. Lane 1, 50 ng of purified CcpA of B. megaterium; lane 2, 0.2 OD600 equivalents of protein extract of L. pentosus LPE4 (ccpA mutant); lanes 3 and 4, 0.2 OD600 equivalents of protein extract of L. pentosus MD363 grown on M medium with 50 mM glucose and 50 mM xylose, respectively.
FIG. 5
Comparison of ccpA regions of L. pentosus, L. delbrueckii subsp. lactis (40, 41), L. casei (; C. Esteban and G. Pérez-Martínez, unpublished data), Streptococcus mutans (37), Lactococcus lactis (accession no. AF106673) (3) (A); B. megaterium (15), B. subtilis (2, 11, 13) (B); and Staphylococcus xylosus (6) (C). Orientations of genes are indicated by arrows. Potential transcriptional termination structures (T) and cre motifs are indicated.
Similar articles
- The functional ccpA gene is required for carbon catabolite repression in Lactobacillus plantarum.
Muscariello L, Marasco R, De Felice M, Sacco M. Muscariello L, et al. Appl Environ Microbiol. 2001 Jul;67(7):2903-7. doi: 10.1128/AEM.67.7.2903-2907.2001. Appl Environ Microbiol. 2001. PMID: 11425700 Free PMC article. - Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus.
Egeter O, Brückner R. Egeter O, et al. Mol Microbiol. 1996 Aug;21(4):739-49. doi: 10.1046/j.1365-2958.1996.301398.x. Mol Microbiol. 1996. PMID: 8878037 - Cloning, sequencing and characterization of the ccpA gene from Enterococcus faecalis.
Leboeuf C, Auffray Y, Hartke A. Leboeuf C, et al. Int J Food Microbiol. 2000 Apr 10;55(1-3):109-13. doi: 10.1016/s0168-1605(00)00185-9. Int J Food Microbiol. 2000. PMID: 10791727 Review. - Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus.
Jankovic I, Brückner R. Jankovic I, et al. J Mol Microbiol Biotechnol. 2002 May;4(3):309-14. J Mol Microbiol Biotechnol. 2002. PMID: 11931563 Review.
Cited by
- Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis.
Asanuma N, Yoshii T, Hino T. Asanuma N, et al. Appl Environ Microbiol. 2004 Sep;70(9):5244-51. doi: 10.1128/AEM.70.9.5244-5251.2004. Appl Environ Microbiol. 2004. PMID: 15345406 Free PMC article. - Salt stress proteins induced in Listeria monocytogenes.
Duché O, Trémoulet F, Glaser P, Labadie J. Duché O, et al. Appl Environ Microbiol. 2002 Apr;68(4):1491-8. doi: 10.1128/AEM.68.4.1491-1498.2002. Appl Environ Microbiol. 2002. PMID: 11916660 Free PMC article. - Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363.
Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Zomer AL, et al. J Bacteriol. 2007 Feb;189(4):1366-81. doi: 10.1128/JB.01013-06. Epub 2006 Oct 6. J Bacteriol. 2007. PMID: 17028270 Free PMC article. - Genetic Elements Orchestrating Lactobacillus crispatus Glycogen Metabolism in the Vagina.
Hertzberger R, May A, Kramer G, van Vondelen I, Molenaar D, Kort R. Hertzberger R, et al. Int J Mol Sci. 2022 May 17;23(10):5590. doi: 10.3390/ijms23105590. Int J Mol Sci. 2022. PMID: 35628398 Free PMC article.
References
- Bolotin A, Khazak V, Stoynova N, Ratmanova K, Yomantas Y, Kozlov Y. Identical amino acid sequence of the aroA(G) gene products of Bacillus subtilis 168 and B. subtilis Marburg strain. Microbiology. 1995;141:2219–2222. - PubMed
- Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. - PubMed
- Davison S P, Santangelo J D, Reid S J, Woods D R. A Clostridium acetobutylicum regulator gene (regA) affecting amylase production in Bacillus subtilis. Microbiology. 1995;141:989–996. - PubMed
- Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous