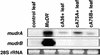The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S -driven MURA cDNA in transgenic maize - PubMed (original) (raw)
The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S -driven MURA cDNA in transgenic maize
M N Raizada et al. Plant Cell. 2000 Jan.
Abstract
The MuDR element responsible for Mutator activities in maize encodes two genes, mudrA and mudrB. Each encodes multiple transcripts hypothesized to regulate, directly or indirectly, the unique late timing and switch in transposition mechanism during maize development. mudrA, which encodes the MURA transposase, is unstable in bacterial plasmids, a technical problem solved by using phage M13 as a vector to prepare DNA for biolistic transformation. In transgenic maize, a single 2.7-kb mudrA cDNA predicted to encode an 823-amino acid protein is sufficient to catalyze late somatic excisions, despite removal of the native promoter, alternative transcription start sites, known introns, polymorphic 5' and 3' untranslated sequences, and the mudrB gene. These results suggest that post-translational regulation confers Mu excision timing. The transgene is active in lines containing silencing MuDR elements. This suggests that endogenous MuDR transposons do not measurably immunize the host against expression of a homologous transgene.
Figures
Figure 1.
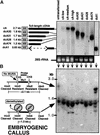
Structure of MuDR, Endogenous mudrA and mudrB Transcripts, the CaMV 35S–mudrA Construct in cA+ Transgenic Maize Lines, and the Probes Used for RNA Gel Blots. (A) Structure of an endogenous MuDR element. The element has two open reading frames, termed mudrA and mudrB, encoded in antiparallel orientation. The intergenic region between the two genes is composed of diverse short repetitive elements. The promoters are located within the ∼215-bp TIRs. The mudrA region with high similarity to bacterial transposases is shown in white. The DNA probes for RNA analysis in this study are located above the element. Numbering is according to Hershberger et al. (1991). (B) The diversity of endogenous mudrA and mudrB transcripts in active Mutator seedlings (Hershberger et al., 1995). Intron sequences shown in solid black are in-frame with exons. Alternative mudrA transcription initiation sites (+169 and +252) produce transcripts with a short or long 5′ leader sequence. aa, amino acids. (C) The structure of the CaMV 35S–mudrA cDNA in M13 transformed into maize to make cA lines. In construct phMR53, the native promoter, alternative start sites, 5′ UTR, and introns were removed. The CaMV 35S promoter and 130-bp leader sequences were substituted. The mudrA 3′ UTR (polymorphic region) was truncated and fused to the nopaline synthase (nos) terminator.
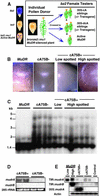
Figure 2.
Test for Interaction of CaMV 35S–mudrA_—Encoded Full-Length and Truncated Proteins with the MURA DNA Binding Sites of Mu1 and Mu2 TIRs in Embryogenic Callus. (A) RNA gel blot hybridization analysis of mudrA transgene transcripts in maize leaves from T0 plants. Lines tested correspond to plants regenerated from each callus line shown in (B). Line dcA1 was probed with the mudrA+B (BX1.0) probe (Figure 1A), which hybridizes with the 3′ end of mudrA. All other lanes were probed with the 1.3-kb 5′ mudrA probe (Figure 1A). The estimated sizes of the truncated transgenes, based on RNA gel blot analysis, are shown at left. At bottom is the ethidium bromide–stained formaldehyde agarose gel indicating the 28S rRNA. (B) At left is a model to explain the effect of MURA binding on the methylation status of HinfI sites in the TIRs of Mu1 and Mu2. MURA binds to an ∼30-bp region overlapping the HinfI site in each TIR (Benito and Walbot, 1997). At right is a DNA gel blot of callus DNA digested with methylation-sensitive HinfI and hybridized with probe pA/B5, which recognizes both Mu1 and Mu2. DNA was prepared from calli 10 to 12 weeks after stable transformation with the CaMV 35S–_mudrA vector.
Figure 3.
RNA Gel Blot Hybridization Analysis of MuDR and CaMV 35S–mudrA Full-Length cDNA Transgenic Lines. Control total RNA is from a leaf of a standard inbred tester. MuDR RNA is from an immature ear of an active, high-copy MuDR line. The transgene samples are from mature leaves of T0 plants. Both mudrA and mudrB are from the same blot, probed with the cross-hybridizing mudrA+B (BX1.0) probe. No mudrB transcript was detected in any cA lines. The 28S rRNA panel is from the ethidium bromide–stained agarose gel.
Figure 4.
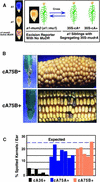
Genetic Test of the Ability of CaMV 35S–mudrA to Catalyze Excisions of Mu1 Elements at the a1 Locus in the Absence of Intact MuDR Elements. (A) At left is a diagram demonstrating the expected aleurone phenotypes of different A1 genotypes. At right is the genetic experiment in which sibling plants carrying the a1-mum2 excision reporter were crossed to or by transgenic T2 generation plants segregating 1:1 for the transgene. cA+ indicates herbicide resistance and cA− indicates herbicide sensitivity. (B) At left are the leaf phenotypes of transgenic line cA75B parents 5 to 7 days after application of Basta herbicide. At right are the phenotypes of progeny of crosses between a1-mum2 and +/− transgene lines. The ears are the T4 generation. (C) Percentage of spotted kernels per ear of cA+ CaMV 35S–mudrA lines. All kernels have one copy of a1-mum2 and one copy of the CaMV 35S–mudrA locus; both genes were transmitted through pollen in crosses onto a1 tester ears. Twenty-five percent of kernels were expected to be spotted. Each bar represents an individual ear.
Figure 5.
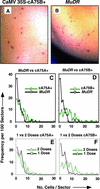
Comparative Analysis of the Developmental Timing of Excision from Aleurone Cells of Mu1 from a1-mum2 in MuDR and CaMV 35S–mudrA Lines. (A) cA75B+ aleurone. Most of the excisions shown consist of single cells. (B) One-copy MuDR aleurone. Most sectors consist of one or two cells. (C) and (D) Sector size distribution comparison. One hundred randomly chosen sectors were scored per line. The MuDR source was selfed, and kernels contain one to three copies of MuDR plus one to three copies of a1-mum2. The cA75+ line contains two doses of CaMV 35S–mudrA and two to three copies of a1-mum2. The MuDR data in both (C) and (D) are the same. The MuDR kernel had one additional 48-cell sector (not shown on graph). (E) and (F) Sector size distribution as a function of CaMV 35S–mudrA transgene locus dose. The two-dose data are from (C) and (D), respectively. The one-dose kernels contain one dose of CaMV 35S–mudrA and two to three copies of a1-mum2. In the two-dose kernels, the transgene had been transmitted through the female. Single-dose kernel transgenes had been transmitted through male germinal cells.
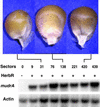
Figure 6.
Aleurone Mu1 Excision Frequency Compared with mudrA Transcript Abundance. All kernels were from a single cA75B+ ear of the T3 generation, and they are ranked by the number of excision sectors in the aleurone, from zero at left to 439 at right. Seedlings were scored for herbicide resistance, and RNA was isolated from the tips of leaf 4. The same RNA gel blot was probed with the 1.3-kb mudrA probe and subsequently with a maize actin probe used as an RNA loading control.
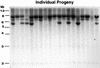
Figure 7.
DNA Gel Blot Screen for Mu1 and Mu2 Germinal Insertions in the Progeny of CaMV 35S–mudrA Plants. Line cA75B+ progeny were used, and all kernels were spotted. DNA was digested with HindIII and probed with the Mu1/_Mu2_–specific probe pA/B5. Only parental fragments, visualized as segregating bands in the progeny, are seen. DNA gel migration size standards are indicated at left.
Figure 8.
Stability of the CaMV 35S–mudrA Transgene in the Presence of Multiple Silencing MuDR Elements. (A) At left is a cartoon of aleurone genotypes and phenotypes at the Bz2 locus. At right is a schematic of the genetic experiment. (B) Kernel progeny showing reactivation of excisions at bz::mu1 from a previously silenced MuDR line. Pollen from a silenced bz2::mu1 individual was crossed onto bz2 testers containing either high-copy active MuDR or the cA75B+ transgene. The cA75B+/− kernels are siblings from the same ear. (C) DNA gel blot analysis of the methylation status at HinfI sites within the MURA binding sites in the TIRs of Mu1 and Mu2 elements (see Figure 2 for assay description). Kernels pictured in (B) were planted; DNA from leaf 4 was digested with HinfI, and DNA gel blots were probed with the Mu1/_Mu2_–specific probe pA/B5. DNA gel migration size standards are indicated at left. (D) RNA gel blot analysis of immature ear tissue from plants derived from cA75B+/− kernels shown in (B) and analyzed in (C). The _mudrA_- or _mudrB-specific probes were hybridized against 15 μg of total RNA. Each lane represents ear RNA pooled from two plants. (E) RT-PCR analysis of cA75B+/− immature ears. Samples are from the plants analyzed in (B) through (D). This analysis was designed to detect the presence of reactivated mudrB (TIR:mudrB) transcripts from the silenced MuDR elements and to distinguish between mudrA transcripts originating from the CaMV 35S–_mudrA transgene or from the silenced MuDR (TIR:mudrA) elements. For RT-PCR, we used promoter-specific 5′ PCR primers. PCR products were DNA gel blotted and probed with the _mudrA_-, _mudrB_-, or actin-specfic probes. The MuDR sample is from a high-copy MuDR line. The control is a non-MuDR tester. Actin primers were included during RT-PCR as an internal control.
Similar articles
- The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize.
Woodhouse MR, Freeling M, Lisch D. Woodhouse MR, et al. Genetics. 2006 Jan;172(1):579-92. doi: 10.1534/genetics.105.051383. Epub 2005 Oct 11. Genetics. 2006. PMID: 16219782 Free PMC article. - Deletion derivatives of the MuDR regulatory transposon of maize encode antisense transcripts but are not dominant-negative regulators of mutator activities.
Kim SH, Walbot V. Kim SH, et al. Plant Cell. 2003 Oct;15(10):2430-47. doi: 10.1105/tpc.014605. Epub 2003 Sep 24. Plant Cell. 2003. PMID: 14508005 Free PMC article. - The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs.
Raizada MN, Benito MI, Walbot V. Raizada MN, et al. Plant J. 2001 Jan;25(1):79-91. doi: 10.1046/j.1365-313x.2001.00939.x. Plant J. 2001. PMID: 11169184 - Regulation of mutator activities in maize.
Walbot V, Britt AB, Luehrsen K, McLaughlin M, Warren C. Walbot V, et al. Basic Life Sci. 1988;47:121-35. doi: 10.1007/978-1-4684-5550-2_9. Basic Life Sci. 1988. PMID: 2845910 Review. - Movers and shakers: maize transposons as tools for analyzing other plant genomes.
Osborne BI, Baker B. Osborne BI, et al. Curr Opin Cell Biol. 1995 Jun;7(3):406-13. doi: 10.1016/0955-0674(95)80097-2. Curr Opin Cell Biol. 1995. PMID: 7662372 Review.
Cited by
- Independent evolution of transposase and TIRs facilitated by recombination between Mutator transposons from divergent clades in maize.
Hunter CT, McCarty DR, Koch KE. Hunter CT, et al. Proc Natl Acad Sci U S A. 2023 Aug;120(31):e2305298120. doi: 10.1073/pnas.2305298120. Epub 2023 Jul 25. Proc Natl Acad Sci U S A. 2023. PMID: 37490540 Free PMC article. - A Novel Mutator-Like Transposable Elements With Unusual Structure and Recent Transpositions in Barley (Hordeum vulgare).
Gao D, Caspersen AM, Hu G, Bockelman HE, Chen X. Gao D, et al. Front Plant Sci. 2022 May 23;13:904619. doi: 10.3389/fpls.2022.904619. eCollection 2022. Front Plant Sci. 2022. PMID: 35677233 Free PMC article. - Transposition of a rice Mutator-like element in the yeast Saccharomyces cerevisiae.
Zhao D, Ferguson A, Jiang N. Zhao D, et al. Plant Cell. 2015 Jan;27(1):132-48. doi: 10.1105/tpc.114.128488. Epub 2015 Jan 13. Plant Cell. 2015. PMID: 25587002 Free PMC article. - Phenotype to genotype using forward-genetic Mu-seq for identification and functional classification of maize mutants.
Hunter CT, Suzuki M, Saunders J, Wu S, Tasi A, McCarty DR, Koch KE. Hunter CT, et al. Front Plant Sci. 2014 Jan 7;4:545. doi: 10.3389/fpls.2013.00545. eCollection 2014 Jan 7. Front Plant Sci. 2014. PMID: 24432026 Free PMC article. - Identification of an active Mutator-like element (MULE) in rice (Oryza sativa).
Gao D. Gao D. Mol Genet Genomics. 2012 Mar;287(3):261-71. doi: 10.1007/s00438-012-0676-x. Epub 2012 Jan 25. Mol Genet Genomics. 2012. PMID: 22274888
References
- Armstrong, C.L. (1994). Regeneration of plants from somatic cell cultures: Applications for in vitro genetic manipulation. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 663–671.
- Armstrong, C.L., and Green, C.E. (1985). Establishment and maintenance of friable, embryogenic maize callus and the involvement of l-proline. Planta 164 207–214 - PubMed
- Benito, M.-I., and Walbot, V. (1994). The terminal inverted repeat sequences of MuDR are functionally active promoters in maize cells. Maydica 39 255–264.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials