Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways - PubMed (original) (raw)
Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways
C V Ramana et al. EMBO J. 2000.
Abstract
Interferons (IFNs) inhibit cell growth in a Stat1-dependent fashion that involves regulation of c-myc expression. IFN-gamma suppresses c-myc in wild-type mouse embryo fibroblasts, but not in Stat1-null cells, where IFNs induce c-myc mRNA rapidly and transiently, thus revealing a novel signaling pathway. Both tyrosine and serine phosphorylation of Stat1 are required for suppression. Induced expression of c-myc is likely to contribute to the proliferation of Stat1-null cells in response to IFNs. IFNs also suppress platelet-derived growth factor (PDGF)-induced c-myc expression in wild-type but not in Stat1-null cells. A gamma-activated sequence element in the promoter is necessary but not sufficient to suppress c-myc expression in wild-type cells. In PKR-null cells, the phosphorylation of Stat1 on Ser727 and transactivation are both defective, and c-myc mRNA is induced, not suppressed, in response to IFN-gamma. A role for Raf-1 in the Stat1-independent pathway is revealed by studies with geldanamycin, an HSP90-specific inhibitor, and by expression of a mutant of p50(cdc37) that is unable to recruit HSP90 to the Raf-1 complex. Both agents abrogated the IFN-gamma-dependent induction of c-myc expression in Stat1-null cells.
Figures
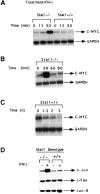
Fig. 1. c–myc mRNA expression in response to IFN–γ in Stat1-null and wild-type MEFs. (A) Subconfluent, serum-starved MEFs were either untreated or treated with 1000 IU/ml of murine IFN–γ for 15 or 30 min. c–myc and GAPDH mRNA levels were analyzed by Northern blotting. (B) Stat1-null cells were treated with murine IFN–γ (1000 IU/ml). c–myc and GAPDH mRNA levels were determined as above. (C) Wild-type cells were treated with murine IFN–γ (1000 IU/ml). c–myc and GAPDH mRNA levels were determined as above. (D) Subconfluent, serum-starved fibroblasts were either untreated or treated with 1000 IU/ml of murine IFN–γ for 30 min. Northern blot analyses were conducted with the probes indicated.
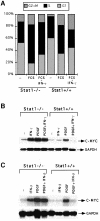
Fig. 2. Effects of growth factor and IFN treatment on c–myc regulation. (A) MEFs were grown to 20% confluence in DMEM with 10% FCS. The cells were serum-starved in DMEM with 0.1% FCS for 48 h. Cells were either untreated (–) or treated with 10% FCS, alone (FCS) or with 1000 IU/ml of murine IFN–γ (FCS+IFN–γ). Twenty-four hours later, the cells were stained with propidium iodide and the DNA content was analyzed by flow cytometry. The percentage of cells in the G1, S and G2+M parts of the cell cycle are indicated in each histogram. (B) Stat1-null or wild-type MEFs were either untreated or treated for 30 min with 1000 IU/ml of IFN–γ alone, 200 ng/ml of PDGF alone, or PDGF plus IFN–γ. Northern transfers were hybridized with c–myc or GAPDH probes. (C) Stat1-null or wild-type MEFs were either untreated or treated for 30 min with 1000 IU/ml of IFN–β alone, 200 ng/ml of PDGF alone, or PDGF plus IFN–β. Northern transfers were hybridized with c–myc or GAPDH probes.
Fig. 3. Regulation of c–myc but not p21_waf1_ expression by IFN–γ in 2fTGH and U3A cells. (A) Subconfluent, serum-starved cells were treated with 1500 IU/ml of human IFN–γ for 60 to 120 min. Northern transfers were hybridized with the probes indicated. (B) Cells were either untreated or treated with 1500 IU/ml of human IFN–γ for 3 or 6 h. p21_waf1_ levels were determined by Western blot analysis.
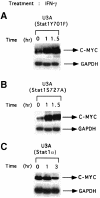
Fig. 4. Stat1 domains required to suppress expression of c–myc mRNA. U3A cells reconstituted with a Stat1 tyrosine (U3A Stat1 Y701F) or serine (U3A Stat1 S727A) phosphorylation site mutant or with wild-type Stat1α (Kumar et al., 1997b; see for expression levels) were treated with 1500 IU/ml of human IFN–γ. c–myc and GAPDH RNA levels were determined by Northern blot analysis.
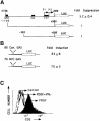
Fig. 5. Identification of a GAS element in the c–myc promoter that is necessary but not sufficient for suppression. (A) The full-length 1.7 kb c–myc promoter or two deletion constructs were linked to luciferase and transfected transiently into 2fTGH cells. Luciferase activity was determined with or without IFN–γ treatment (1500 IU/ml for 6 h). The data shown represent duplicate experiments in three independent trials (standard deviations). (B) Constructs containing eight copies of the consensus GAS (TTCTCGGAA) or seven copies of the c–myc GAS (TTCTGGGAA) linked to luciferase were transfected transiently into 2fTGH cells. Luciferase activity was determined with or without IFN–γ treatment (1500 IU/ml for 6 h). Data are presented from three independent experiments, with standard deviations. (C) NIH 3T3 cells stably transfected with the 1.7 kb c–myc promoter linked to the cell-surface protein cd2 were serum-starved and treated with PDGF alone (200 ng/ml), PDGF plus IFN–γ (1000 IU/ml) or were untreated. cd2 expression was determined by FACScan analysis.
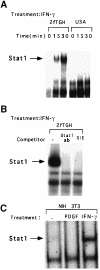
Fig. 6. Binding of Stat1 to the c–myc GAS. (A) EMSAs were performed with whole-cell extracts prepared from U3A or 2fTGH cells, not treated or treated for 15 or 30 min with 1000 IU/ml of IFN–γ. (B) EMSAs were performed with whole-cell extracts from 2fTGH cells treated with 1000 IU/ml of IFN–γ. The extracts were pre-incubated with anti-Stat1 or with a 100–fold molar excess of the unlabeled SIE (m67) GAS. (C) EMSAs were performed with whole-cell extracts from NIH 3T3 cells, treated for 30 min with 200 ng/ml of PDGF, 1000 IU/ml of murine IFN–γ, or not treated.
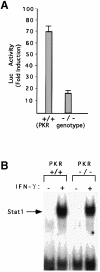
Fig. 7. Defective Stat1 activation and serine phosphorylation in PKR-null cells. (A) 8× con GAS linked to luciferase was transiently transfected into wild-type or PKR-null MEFs. Luciferase activity was determined with or without murine IFN–γ (1000 IU/ml) for 6 h. Results are presented with standard deviations from three independent experiments. (B) Whole-cell extracts were prepared from serum-starved wild-type or PKR-null cells, with or without treatment with 1000 IU/ml of murine IFN–γ. Stat1 binding to the SIE (m67) GAS was determined by EMSA. (C) Extracts of serum-starved cells, either untreated or treated with IFN–γ (1000 IU/ml for 20 min) were immunoprecipitated with anti-Stat1. The transfer was probed first with an antibody specific for a Stat1 peptide that includes phosphorylated Ser727 and then reprobed with anti-Stat1. (D) RNA from MEFs untreated or treated with murine IFN–γ (1000 IU/ml) was analyzed by the Northern procedure.
Fig. 7. Defective Stat1 activation and serine phosphorylation in PKR-null cells. (A) 8× con GAS linked to luciferase was transiently transfected into wild-type or PKR-null MEFs. Luciferase activity was determined with or without murine IFN–γ (1000 IU/ml) for 6 h. Results are presented with standard deviations from three independent experiments. (B) Whole-cell extracts were prepared from serum-starved wild-type or PKR-null cells, with or without treatment with 1000 IU/ml of murine IFN–γ. Stat1 binding to the SIE (m67) GAS was determined by EMSA. (C) Extracts of serum-starved cells, either untreated or treated with IFN–γ (1000 IU/ml for 20 min) were immunoprecipitated with anti-Stat1. The transfer was probed first with an antibody specific for a Stat1 peptide that includes phosphorylated Ser727 and then reprobed with anti-Stat1. (D) RNA from MEFs untreated or treated with murine IFN–γ (1000 IU/ml) was analyzed by the Northern procedure.
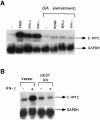
Fig. 8. Pre-treatment with geldanamycin or dominant-negative p50_cdc37_ abrogates the induction of c–myc by IFN–γ. (A) Stat1-null MEFs were serum-starved and either untreated or pre-treated with geldanamycin (GA, 2 μg/ml) for 5 h. The cells were unstimulated or stimulated with IFN–γ, IFN–β or PDGF for 30 min. c–myc and GAPDH mRNA levels were determined by Northern blot analysis. (B) Stat1-null MEFs were transiently transfected with vector alone or vector encoding dominant-negative p50_cdc37_. The transfected cells were serum-starved for 24 h and either unstimulated or stimulated with IFN–γ for 30 min. c–myc and GAPDH RNA levels were determined as above.
Similar articles
- Stat1-independent regulation of gene expression in response to IFN-gamma.
Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Ramana CV, et al. Proc Natl Acad Sci U S A. 2001 Jun 5;98(12):6674-9. doi: 10.1073/pnas.111164198. Proc Natl Acad Sci U S A. 2001. PMID: 11390994 Free PMC article. - A PI-3 kinase-dependent, Stat1-independent signaling pathway regulates interferon-stimulated monocyte adhesion.
Navarro A, Anand-Apte B, Tanabe Y, Feldman G, Larner AC. Navarro A, et al. J Leukoc Biol. 2003 Apr;73(4):540-5. doi: 10.1189/jlb.1002508. J Leukoc Biol. 2003. PMID: 12660229 - Stat1-independent induction of SOCS-3 by interferon-gamma is mediated by sustained activation of Stat3 in mouse embryonic fibroblasts.
Ramana CV, Kumar A, Enelow R. Ramana CV, et al. Biochem Biophys Res Commun. 2005 Feb 18;327(3):727-33. doi: 10.1016/j.bbrc.2004.12.074. Biochem Biophys Res Commun. 2005. PMID: 15649407 - Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling.
Ramana CV, Gil MP, Schreiber RD, Stark GR. Ramana CV, et al. Trends Immunol. 2002 Feb;23(2):96-101. doi: 10.1016/s1471-4906(01)02118-4. Trends Immunol. 2002. PMID: 11929133 Review. - Regulation of Myc: Max complex formation and its potential role in cell proliferation.
Blackwood EM, Eisenman RN. Blackwood EM, et al. Tohoku J Exp Med. 1992 Oct;168(2):195-202. doi: 10.1620/tjem.168.195. Tohoku J Exp Med. 1992. PMID: 1306304 Review.
Cited by
- Human CD34+-derived plasmacytoid dendritic cells as surrogates for primary pDCs and potential cancer immunotherapy.
Fiore G, Weckwarth W, Paetzold K, Albertí Servera L, Gies M, Rosenhauer J, Antoniolli M, Nassiri S, Schmeing S, Dettling S, Soni B, Majety M, Krug AB, Hoves S, Wolf MJ. Fiore G, et al. Front Immunol. 2024 Nov 7;15:1433119. doi: 10.3389/fimmu.2024.1433119. eCollection 2024. Front Immunol. 2024. PMID: 39575246 Free PMC article. - Cold and hot tumors: from molecular mechanisms to targeted therapy.
Wu B, Zhang B, Li B, Wu H, Jiang M. Wu B, et al. Signal Transduct Target Ther. 2024 Oct 18;9(1):274. doi: 10.1038/s41392-024-01979-x. Signal Transduct Target Ther. 2024. PMID: 39420203 Free PMC article. Review. - Restriction and evasion: a review of IFNγ-mediated cell-autonomous defense pathways during genital Chlamydia infection.
Reitano JR, Coers J. Reitano JR, et al. Pathog Dis. 2024 Feb 7;82:ftae019. doi: 10.1093/femspd/ftae019. Pathog Dis. 2024. PMID: 39210512 Free PMC article. Review. - Triptolide, a Cancer Cell Proliferation Inhibitor, Causes Zebrafish Muscle Defects by Regulating Notch and STAT3 Signaling Pathways.
Lee B, Park Y, Lee Y, Kwon S, Shim J. Lee B, et al. Int J Mol Sci. 2024 Apr 25;25(9):4675. doi: 10.3390/ijms25094675. Int J Mol Sci. 2024. PMID: 38731894 Free PMC article. - Deregulated protein homeostasis constrains fetal hematopoietic stem cell pool expansion in Fanconi anemia.
Kovuru N, Mochizuki-Kashio M, Menna T, Jeffrey G, Hong Y, Me Yoon Y, Zhang Z, Kurre P. Kovuru N, et al. Nat Commun. 2024 Feb 29;15(1):1852. doi: 10.1038/s41467-024-46159-1. Nat Commun. 2024. PMID: 38424108 Free PMC article.
References
- Balkwill F. and Taylor-Papadimitriou, J. (1978) Interferon affects both G1 and S+G2 in cells stimulated from quiescence to growth. Nature, 274, 798–801. - PubMed
- Bao J. and Zervos, A.S. (1996) Isolation and characterization of Nmi, a novel partner of Myc proteins. Oncogene, 12, 2171–2176. - PubMed
- Bennett M.R., Evan, G.I. and Newby, A.C. (1994) Deregulated expression of the c–myc oncogene abolishes inhibition of proliferation of rat vascular smooth muscle cells by serum reduction, interferon-γ, heparin, and cyclic nucleotide analogues and induces apoptosis. Circ. Res., 74, 525–536. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous