Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53 - PubMed (original) (raw)
. 2000 Feb 1;14(3):278-88.
Affiliations
- PMID: 10673500
- PMCID: PMC316357
Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53
N H Chehab et al. Genes Dev. 2000.
Abstract
Chk2/hcds1, the human homolog of the Saccharomyces cerevisiae RAD53/SPK1 and Schizosaccharomyces pombe cds1 DNA damage checkpoint genes, encodes a protein kinase that is post-translationally modified after DNA damage. Like its yeast homologs, the Chk2/hCds1 protein phosphorylates Cdc25C in vitro, suggesting that it arrests cells in G(2) in response to DNA damage. We expressed Chk2/hCds1 in human cells and analyzed their cell cycle profile. Wild-type, but not catalytically inactive, Chk2/hCds1 led to G(1) arrest after DNA damage. The arrest was inhibited by cotransfection of a dominant-negative p53 mutant, indicating that Chk2/hCds1 acted upstream of p53. In vitro, Chk2/hCds1 phosphorylated p53 on Ser-20 and dissociated preformed complexes of p53 with Mdm2, a protein that targets p53 for degradation. In vivo, ectopic expression of wild-type Chk2/hCds1 led to increased p53 stabilization after DNA damage, whereas expression of a dominant-negative Chk2/hCds1 mutant abrogated both phosphorylation of p53 on Ser-20 and p53 stabilization. Thus, in response to DNA damage, Chk2/hCds1 stabilizes the p53 tumor suppressor protein leading to cell cycle arrest in G(1).
Figures
Figure 1
Chk2/hCds1 phosphorylates p53 on Ser-20 in vitro and dissociates preformed p53/Mdm2 complexes. (A) Recombinant Chk2/hCds1 was incubated with [32P]ATP in the presence or absence of recombinant full-length p53. The reaction products were resolved by denaturing gel electrophoresis and visualized by autoradiography. (B) Full-length wild-type (wt) p53 or p53 with substitution of Ser-20 with Ala (A20) were incubated with Chk2/hCds1. Phosphorylation of p53 on Ser-20 (S20p) was monitored by immunoprecipitation of the reaction products with AbS20p, an antibody specific for p53 phosphorylated on Ser-20, followed by immunoblotting with antibody DO7, which recognizes p53 irrespective of its phosphorylation state. p53 protein levels were monitored by immunoblotting with DO7. (C) p53 phosphorylated by Chk2/hCds1 does not interact with Mdm2. Full-length p53 was incubated with Chk2/hCds1. One-half of the sample was stored on ice (input sample) and the other half was incubated with beads coated with GST–Mdm2(1–125). After washing the beads to remove unbound p53, the p53 protein bound to Mdm2 was eluted (Mdm2-bound sample). The fraction of p53 phosphorylated on Ser-20 in the input and Mdm2-bound samples was determined by comparing the amounts of p53 immunoprecipitated with antibodies AbS20p (p53 phosphorylated on Ser-20; S20p) and DO7 (p53 phosphorylated and nonphosphorylated on Ser-20; S20+S20p). (D) Chk2/hCds1 disrupts preformed p53/Mdm2 complexes. Complexes of GST–Mdm2 with wild-type p53 or p53A20 bound to glutathione beads were incubated with Chk2/hCds1. p53 that remained bound to Mdm2 after incubation of the beads with Chk2/hCds1 was resolved by denaturing gel electrophoresis and detected by immunoblotting with antibody DO7.
Figure 2
Time course of p53 stabilization and Chk2/hCds1 activation in U2–OS cells exposed to IR or UV light. Extracts from cells exposed to DNA damage were resolved by denaturing gel electrophoresis and immunoblotted for p53 and Flag-tagged Chk2/hCds1. Chk2/hCds1 activated in response to DNA damage exhibits a mobility shift on denaturing gels.
Figure 3
ATM dependence of p53 phosphorylation on Ser-20 in response to IR, but not UV light. Primary fibroblasts from a normal individual (AG1522) or from A-T patients (AT1BR and AT5BI) were exposed to IR or UV light. Phosphorylation of p53 on Ser-20 (S20p) was monitored with the phosphate-specific antibody AbS20p. p53* indicates that to facilitate analysis of the p53 phosphorylation state, the amounts of extract used in each lane of the top and bottom panels were adjusted so that all lanes had equal levels of total p53 (bottom; immunoblotting with antibody DO7).
Figure 4
Dominant-negative Chk2/hCds1 mutants inhibit DNA damage-induced p53 stabilization (A) and Ser-20 phosphorylation (B). U2–OS cells were transiently transfected with plasmids expressing HA-tagged p53, the indicated Flag-tagged Chk2/hCds1 protein and GFP, as a marker of transfection efficiency. The cells were exposed to IR or UV light and cell extracts were prepared 2 and 16 hr later, respectively. (A) HA-tagged p53, Flag-tagged Chk2/hCds1, and GFP were resolved by denaturing gel electrophoresis and detected by immunoblotting. Equal levels of total protein were loaded in each lane to monitor the increase in p53 protein levels. (B) Phosphorylation of HA-tagged p53 on Ser-20 was monitored by immunoprecipitation with AbS20p, followed by immunoblotting with an antibody that recognizes the HA tag. The interaction of HA-tagged p53 with 14-3-3 proteins was monitored by immunoprecipitation with an antibody that recognizes 14-3-3 and detection of the coprecipitated HA-tagged p53 by immunoblotting. p53* indicates that to facilitate analysis of the p53 phosphorylation state and interaction with 14-3-3, the amounts of extract used in each lane of the top, middle, and bottom panels were adjusted so that all lanes had equal levels of total p53 (bottom; immunoblotting with an antibody that recognizes HA-tagged p53).
Figure 5
Ectopic expression of Chk2/hCds1 leads to p53 stabilization. (A) U2–OS cells transiently transfected with plasmids expressing Neo or Chk2/hCds1 and GFP, as a marker, were exposed to IR or were mock irradiated. Cell extracts were prepared 2 hr later, resolved by denaturing gel electrophoresis and immunoblotted for endogenous p53 with antibody DO7 and for GFP to monitor transfection efficiency. (B) U2–OS cells transiently transfected with plasmids expressing Neo, Chk2/hCds1, or wild-type p53 were exposed to IR. Two hours later, p53 levels were monitored by immunofluorescence and cell nuclei were visualized by DAPI staining.
Figure 6
Chk2/hCds1 functions as a G1 checkpoint in response to DNA damage. (A) Diagram of Chk2/hCds1 protein and deletion mutants. (SQ) Region that contains the SQ/TQ motifs that are putatively phosphorylated by ATM; (FHA) FHA domain; (KD) kinase domain. (B) Expression of Flag-tagged Chk2/hCds1 proteins in transiently transfected U2–OS cells exposed to IR. Extracts from transfected cells were resolved by denaturing gel electrophoresis and immunoblotted with an antibody that recognizes the Flag tag (p53W248, which does not have a Flag tag, is not recognized by this antibody). (C) Chk2/hCds1 induces cell cycle arrest in G1. U2–OS cells were either nontransfected (−) or transiently transfected with plasmids expressing the indicated Chk2/hCds1 and p53 checkpoint proteins and a plasmid expressing GFP (as a marker). Cell cycle profile was determined 12 hr after exposure of the cells to IR. GFP− cells do not express proteins from the transfected plasmids and serve as negative controls. 2N and 4N DNA contents correspond to cells in G1 and G2/M, respectively.
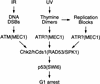
Figure 7
Model for p53 stabilization in response to DNA damage and stalled replication. IR, induces DNA DSBs and activates Chk2/hCds1 in an ATM-dependent manner. Thymine dimers, which are induced by UV light and stalled replication, activate Chk2/hCds1 in an ATM-independent manner. Stabilization of p53 in response to UV light is dependent on ATR and requires DNA replication, suggesting that ATR signals the presence of thymine dimers and/or replication blocks to Chk2/hCds1 (? indicates that it is not known whether ATR responds to thymine dimers or stalled replication or both). In budding yeast, activation of RAD53/SPK1 (the Chk2/hCds1 homolog) is dependent on MEC1 (the ATM and ATR homolog) and leads to phosphorylation of the SWI6 transcription factor and G1 arrest.
Similar articles
- Role of human Cds1 (Chk2) kinase in DNA damage checkpoint and its regulation by p53.
Tominaga K, Morisaki H, Kaneko Y, Fujimoto A, Tanaka T, Ohtsubo M, Hirai M, Okayama H, Ikeda K, Nakanishi M. Tominaga K, et al. J Biol Chem. 1999 Oct 29;274(44):31463-7. doi: 10.1074/jbc.274.44.31463. J Biol Chem. 1999. PMID: 10531348 - The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites.
Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. Shieh SY, et al. Genes Dev. 2000 Feb 1;14(3):289-300. Genes Dev. 2000. PMID: 10673501 Free PMC article. - DNA damage-induced activation of p53 by the checkpoint kinase Chk2.
Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. Hirao A, et al. Science. 2000 Mar 10;287(5459):1824-7. doi: 10.1126/science.287.5459.1824. Science. 2000. PMID: 10710310 - How to activate p53.
Caspari T. Caspari T. Curr Biol. 2000 Apr 20;10(8):R315-7. doi: 10.1016/s0960-9822(00)00439-5. Curr Biol. 2000. PMID: 10801407 Review. - Checking in on Cds1 (Chk2): A checkpoint kinase and tumor suppressor.
McGowan CH. McGowan CH. Bioessays. 2002 Jun;24(6):502-11. doi: 10.1002/bies.10101. Bioessays. 2002. PMID: 12111733 Review.
Cited by
- Systematic transcriptomic analysis and temporal modelling of human fibroblast senescence.
Scanlan RL, Pease L, O'Keefe H, Martinez-Guimera A, Rasmussen L, Wordsworth J, Shanley D. Scanlan RL, et al. Front Aging. 2024 Aug 29;5:1448543. doi: 10.3389/fragi.2024.1448543. eCollection 2024. Front Aging. 2024. PMID: 39267611 Free PMC article. - Protein-truncating and rare missense variants in ATM and CHEK2 and associations with cancer in UK Biobank whole-exome sequence data.
Mukhtar TK, Wilcox N, Dennis J, Yang X, Naven M, Mavaddat N, Perry JRB, Gardner E, Easton DF. Mukhtar TK, et al. J Med Genet. 2024 Oct 23;61(11):1016-1022. doi: 10.1136/jmg-2024-110127. J Med Genet. 2024. PMID: 39209703 Free PMC article. - Mechanisms of DNA Damage Response in Mammalian Oocytes.
Sun F, Sutovsky P, Patterson AL, Balboula AZ. Sun F, et al. Adv Anat Embryol Cell Biol. 2024;238:47-68. doi: 10.1007/978-3-031-55163-5_3. Adv Anat Embryol Cell Biol. 2024. PMID: 39030354 Review. - Single-cell and bulk transcriptional profiling of mouse ovaries reveals novel genes and pathways associated with DNA damage response in oocytes.
Mills M, Emori C, Kumar P, Boucher Z, George J, Bolcun-Filas E. Mills M, et al. bioRxiv [Preprint]. 2024 Feb 4:2024.02.02.578648. doi: 10.1101/2024.02.02.578648. bioRxiv. 2024. PMID: 38352597 Free PMC article. Updated. Preprint. - DNA-Dependent Protein Kinase Inhibitor Peposertib Potentiates the Cytotoxicity of Topoisomerase II Inhibitors in Synovial Sarcoma Models.
Revia S, Budzinska MA, Bogatyrova O, Neumann F, Zimmermann A, Amendt C, Albers J. Revia S, et al. Cancers (Basel). 2023 Dec 30;16(1):189. doi: 10.3390/cancers16010189. Cancers (Basel). 2023. PMID: 38201616 Free PMC article.
References
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, et al. Heterozygous germline hChk2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA09171/CA/NCI NIH HHS/United States
- P01 CA025874/CA/NCI NIH HHS/United States
- CA76367/CA/NCI NIH HHS/United States
- R01 CA076367/CA/NCI NIH HHS/United States
- T32 CA009171/CA/NCI NIH HHS/United States
- CA25874/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous