A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome - PubMed (original) (raw)
A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome
J Hirst et al. J Cell Biol. 2000.
Abstract
We have cloned and characterized members of a novel family of proteins, the GGAs. These proteins contain an NH(2)-terminal VHS domain, one or two coiled-coil domains, and a COOH-terminal domain homologous to the COOH-terminal "ear" domain of gamma-adaptin. However, unlike gamma-adaptin, the GGAs are not associated with clathrin-coated vesicles or with any of the components of the AP-1 complex. GGA1 and GGA2 are also not associated with each other, although they colocalize on perinuclear membranes. Immunogold EM shows that these membranes correspond to trans elements of the Golgi stack and the TGN. GST pulldown experiments indicate that the GGA COOH-terminal domains bind to a subset of the proteins that bind to the gamma-adaptin COOH-terminal domain. In yeast there are two GGA genes. Deleting both of these genes results in missorting of the vacuolar enzyme carboxypeptidase Y, and the cells also have a defective vacuolar morphology phenotype. These results indicate that the function of the GGAs is to facilitate the trafficking of proteins between the TGN and the vacuole, or its mammalian equivalent, the lysosome.
Figures
Figure 1
Sequences, alignments, and domain organization of human GGAs. a, The complete protein sequences of the three mammalian GGAs. GGA1 and GGA2 are 45% identical to each other, and 45 and 35% identical, respectively, to GGA3. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers AF233521 (GGA1), AF233522 (GGA2), and D63876 (GGA3; the GGA3 sequence was entered into the database by Nagase et al. 1995). b, Comparison of the VHS domain of GGA1 with the VHS domains of Hrs, Vps27p, and STAM. GGA1 can be seen to have a typical VHS domain. c, Comparison of the COOH terminus of GGA1 with the COOH termini of the two mammalian γ-adaptin isoforms, γ1 and γ2, and the putative γ-adaptin homologue in S. cerevisiae, Alp4p. All four proteins can be seen to have related COOH-terminal ear domains. d, Schematic diagram of GGA1, GGA2, and GGA3, showing the positions of the VHS domain, the coiled coil domains (CC), the central variable region, and the γ-adaptin ear homology domain.
Figure 3
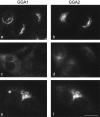
Immunofluorescence localization of GGA1 and GGA2. NRK cells, either nontransfected (a and c) or stably expressing FLAG-tagged GGA2 (b, d, e, and f), were labeled with either an affinity-purified rabbit polyclonal antibody against GGA1 (a, c, and e) or a mouse mAb against the FLAG tag (b, d, and f). Both proteins can be seen to have a punctate distribution in the perinuclear region of the cell (a and b, e and f). The cells shown in c and d were incubated with 100 μg/ml BFA for 2 min at 37°C before fixation. Both proteins have been redistributed to the cytoplasm. The cells in e and f were double-labeled. The two patterns appear to be coincident. Bar: (a–d) 20 μm; (e and f) 14 μm.
Figure 2
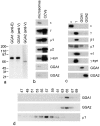
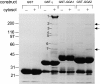
Biochemical characterization of GGA1 and GGA2. a, Western blots of whole HeLa cell extracts probed for GGA1 and GGA2. Two GGA1 antibodies were used, one raised against the COOH-terminal ear domain (anti-E) and one against the central variable region (anti-V). The GGA2 antibody was raised against the variable region. The GGA1 antibodies label a band with an apparent molecular weight of 85 kD, while the GGA2 antibody labels a band with an apparent molecular weight of 67 kD. b, Equal protein loadings of clathrin-coated vesicles purified from rat liver and a crude microsomal fraction from a previous stage in the preparation were subjected to SDS-PAGE, followed by Western blotting. The blot shows that the γ and μ1 subunits of the AP-1 complex and the μ2 subunit of the AP-2 complex, as well as the γ-adaptin binding protein γ-synergin, are strongly enriched in clathrin-coated vesicles, whereas the ε subunit of the AP-4 complex, GGA1, and GGA2 are not. c, Cytosol was prepared from HeLa cells and immunoprecipitated under nondenaturing conditions using affinity-purified antibodies raised against GGA1, GGA2, the AP-4 ε subunit, and the AP-1 γ subunit. Western blots of the immunoprecipitates were probed with antibodies against ε, γ, β1, μ1, σ1, γ-synergin, GGA-1, and GGA-2. The results show that GGA1 and GGA2 do not associate with each other or with the AP-1 complex. c, Pig brain cytosol was fractionated on a Superose 6 column and Western blots of the odd-numbered fractions were probed with antibodies against the μ1 subunit of AP-1, GGA1, and GGA2. AP-1, as marked by μ1, eluted in fractions corresponding to >200 kD, whereas GGA1 and GGA2 eluted in similar positions in fractions corresponding to ∼75 kD.
Figure 4
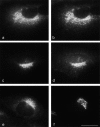
Distribution of GGAs compared with other TGN-associated proteins. a–d, NRK cells stably expressing FLAG-tagged GGA-2 were double-labeled with mouse anti-FLAG antibody (a and c) together with antibodies against the γ-adaptin subunit of the AP-1 adaptor complex (b) and γ-synergin (d). The patterns are similar, but there is more peripheral labeling with the anti–γ-adaptin and anti–γ-synergin antibodies, and the actual patterns of dots are generally distinct. e and f, NRK cells were double-labeled with an affinity-purified polyclonal antibody against GGA1 (e) and an mAb against TGN38 (f). Again, the patterns are similar, although the GGA1 labeling is more punctate than the TGN38 labeling. Bar, 20 μm.
Figure 5
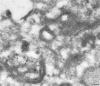
Localization of GGA1 at the electron microscope level. NRK cells were permeabilized by freezing and thawing, then allowed to recruit proteins from pig brain cytosol in the presence of ATP, an ATP-regenerating system, and GTPγS. Frozen thin sections were labeled with an antibody against GGA1, followed by protein A coupled to 15-nm colloidal gold. Labeling can be seen to be associated with the Golgi stack and with tubulovesicular membranes in the Golgi region. Clathrin-coated budding profiles can be seen in the same vicinity (arrowheads), indicating that these membranes correspond to trans-Golgi cisternae and the TGN. Bar, 200 nm.
Figure 6
COOH-terminal domain binding partners. Pig brain cytosol was incubated with GST alone, with a GST fusion protein containing the γ-adaptin ear domain (GST-γ), with a GST fusion protein containing the GGA1 ear domain (GST-GGA1), or with a GST fusion protein containing the GGA2 ear domain (GST-GGA2), followed by glutathione-Sepharose. The samples were subjected to SDS-PAGE and stained with Coomassie blue. The arrows indicate bands that are brought down by all three fusion proteins. The numbered bands were analyzed by MALDI mass spectrometry.
Figure 8
_GGA_-deficient yeast have a vacuolar protein sorting phenotype. a, Yeast cells were pulse-labeled with Promix 35S for 10 min, chased for 30 min, immunoprecipitated for CPY, and subjected to SDS-PAGE. Yeast cells deleted for either GGA gene alone show no defect in CPY processing compared with control wild-type cells. However, cells deleted for both genes (_gga1_Δ/_gga2_Δ) accumulate more of the p2 relative to the m form of CPY, as well as a pseudomature form of CPY. In cells deleted for VPS35 (_vps35_Δ), mainly the p2 form accumulates. b, Wild-type, _gga1_Δ/_gga2_Δ, and _vps35_Δ cells were transformed with either empty vector, yeast GGA1, or a cDNA encoding human GGA2 (hGGA2), and were then assayed for CPY processing as above. The yeast GGA1 gene, but not the mammalian cDNA, restores the wild-type phenotype. c, CPY is secreted from _gga1_Δ/_gga2_Δ cells. Yeast cells were pulse-labeled as above, spheroplasted, and separated into intracellular and extracellular fractions. CPY was immunoprecipitated from both fractions and subjected to SDS-PAGE. The _gga1_Δ/_gga2_Δ strain secretes CPY in the p2 and pseudomature forms. This is in contrast to the wild-type cells, which do not secrete CPY, and the _vps35_Δ cells, which secrete CPY in the p2 form. d, Kinetics of CPY processing. Yeast cells were pulse-labeled with Promix 35S for 3 min, chased for 0–60 min, and then immunoprecipitated for CPY and subjected to SDS-PAGE. _gga1_Δ/_gga2_Δ cells lose the p1 form with wild-type kinetics, but then exhibit slow processing of the p2 form and an accumulation of the pseudomature form(s).
Figure 7
Sequences, alignments, and domain organization of two yeast GGAs. a, The complete protein sequences of yeast Gga1p (Ydr358w) and yeast Gga2p (Yhr108w) aligned with mammalian GGA1. The two yeast sequences are 49% identical to each other, and each is ∼20% identical to each of the three mammalian GGAs. b, Domain organization of Gga1p and Gga2p. Like their mammalian counterparts, the yeast sequences contain a VHS domain, two coiled coil domains (CC), a central variable region, and a γ-adaptin ear homology domain.
Figure 9
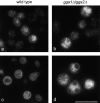
Localization of Vps10p and vacuolar morphology in _gga1_Δ/_gga2_Δ cells. a and b, Yeast cells were transformed with a myc-tagged Vps10p construct, then prepared for immunofluorescence and labeled with a mouse anti-myc antibody. The distribution of Vps10p is similar in the wild-type (a) and _gga1_Δ/_gga2_Δ cells (b), indicating that the GGA genes are not class E VPS genes. b, Cells were pulse-labeled with FM4-64 for 15 min, chased for 60 min at 30°C to label the vacuole, and then immobilized on a microscope slide. In the _gga1_Δ/_gga2_Δ cells (d) the morphology of the vacuole is significantly altered compared with wild-type cells (c), with highly fragmented vacuoles. Scale bar: 10 μm (a and b); 8 μm (c and d).
Similar articles
- GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex.
Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. Dell'Angelica EC, et al. J Cell Biol. 2000 Apr 3;149(1):81-94. doi: 10.1083/jcb.149.1.81. J Cell Biol. 2000. PMID: 10747089 Free PMC article. - GGAs: roles of the different domains and comparison with AP-1 and clathrin.
Hirst J, Lindsay MR, Robinson MS. Hirst J, et al. Mol Biol Cell. 2001 Nov;12(11):3573-88. doi: 10.1091/mbc.12.11.3573. Mol Biol Cell. 2001. PMID: 11694590 Free PMC article. - Adaptor gamma ear homology domain conserved in gamma-adaptin and GGA proteins that interact with gamma-synergin.
Takatsu H, Yoshino K, Nakayama K. Takatsu H, et al. Biochem Biophys Res Commun. 2000 May 19;271(3):719-25. doi: 10.1006/bbrc.2000.2700. Biochem Biophys Res Commun. 2000. PMID: 10814529 - The structure and function of GGAs, the traffic controllers at the TGN sorting crossroads.
Nakayama K, Wakatsuki S. Nakayama K, et al. Cell Struct Funct. 2003 Oct;28(5):431-42. doi: 10.1247/csf.28.431. Cell Struct Funct. 2003. PMID: 14745135 Review. - The GGA proteins: key players in protein sorting at the trans-Golgi network.
Ghosh P, Kornfeld S. Ghosh P, et al. Eur J Cell Biol. 2004 Jul;83(6):257-62. doi: 10.1078/0171-9335-00374. Eur J Cell Biol. 2004. PMID: 15511083 Review.
Cited by
- Regulation of post-Golgi traffic of G protein-coupled receptors.
Wu G. Wu G. Subcell Biochem. 2012;63:83-95. doi: 10.1007/978-94-007-4765-4_5. Subcell Biochem. 2012. PMID: 23161134 Free PMC article. Review. - The Kelch13 compartment contains highly divergent vesicle trafficking proteins in malaria parasites.
Schmidt S, Wichers-Misterek JS, Behrens HM, Birnbaum J, Henshall IG, Dröge J, Jonscher E, Flemming S, Castro-Peña C, Mesén-Ramírez P, Spielmann T. Schmidt S, et al. PLoS Pathog. 2023 Dec 1;19(12):e1011814. doi: 10.1371/journal.ppat.1011814. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38039338 Free PMC article. - Adaptor autoregulation promotes coordinated binding within clathrin coats.
Hung CW, Aoh QL, Joglekar AP, Payne GS, Duncan MC. Hung CW, et al. J Biol Chem. 2012 May 18;287(21):17398-17407. doi: 10.1074/jbc.M112.349035. Epub 2012 Mar 28. J Biol Chem. 2012. PMID: 22457357 Free PMC article. - Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae.
Bonangelino CJ, Chavez EM, Bonifacino JS. Bonangelino CJ, et al. Mol Biol Cell. 2002 Jul;13(7):2486-501. doi: 10.1091/mbc.02-01-0005. Mol Biol Cell. 2002. PMID: 12134085 Free PMC article. - Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B. Reider A, et al. EMBO J. 2009 Oct 21;28(20):3103-16. doi: 10.1038/emboj.2009.248. Epub 2009 Aug 27. EMBO J. 2009. PMID: 19713939 Free PMC article.
References
- Asao H., Sasaki Y., Arita T., Tanaka N., Endo K., Kasai H., Takeshita T., Endo Y., Fujita T., Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 1997;272:32785–32791. - PubMed
- Cowles C.R., Odorizzi G., Payne G.S., Emr S.D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous