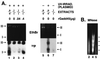p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes - PubMed (original) (raw)
p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes
M L Smith et al. Mol Cell Biol. 2000 May.
Abstract
Human cells lacking functional p53 exhibit a partial deficiency in nucleotide excision repair (NER), the pathway for repair of UV-induced DNA damage. The global genomic repair (GGR) subpathway of NER, but not transcription-coupled repair (TCR), is mainly affected by p53 loss or inactivation. We have utilized mouse embryo fibroblasts (MEFs) lacking p53 genes or downstream effector genes of the p53 pathway, gadd45 (Gadd45a) or p21 (Cdkn1a), as well as MEFs lacking both gadd45 and p21 genes to address the potential contribution of these downstream effectors to p53-associated DNA repair. Loss of p53 or gadd45 had a pronounced effect on GGR, while p21 loss had only a marginal effect, determined by measurements of repair synthesis (unscheduled DNA synthesis), by immunoassays to detect removal of UV photoproducts from genomic DNA, and by assays determining strand-specific removal of CPDs from the mouse dhfr gene. Taken together, the evidence suggests a role for Gadd45, but relatively little role for p21, in DNA repair responses to UV radiation. Recent evidence suggests that Gadd45 binds to UV-damaged chromatin and may affect lesion accessibility. MEFs lacking p53 or gadd45 genes exhibited decreased colony-forming ability after UV radiation and cisplatin compared to wild-type MEFs, indicating their sensitivity to DNA damage. We provide evidence that Gadd45 affects chromatin remodelling of templates concurrent with DNA repair, thus indicating that Gadd45 may participate in the coupling between chromatin assembly and DNA repair.
Figures
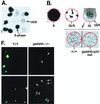
FIG. 1
DNA repair deficiency in cells lacking p53 or p53 effector genes, gadd45 and p21, measured by UDS. (A) Illustration of UDS technique. A low-magnification (×20) field of cells irradiated with 20 J of UV radiation m−2 is shown. Nuclei were made visible by the incorporation of [3H]thymidine during either replicative or repair DNA synthesis. S-phase nuclei appear black upon the photographic emulsion, while non-S-phase cells exhibit UDS. The _p21_−/− and _gadd45/p21_-null lines did exhibit a higher S-phase fraction (50%) compared to wild-type or _gadd45_−/− lines (15 to 20%). (B) Distinctions between replicative DNA synthesis and repair synthesis (UDS). S-phase or G1/S transitional nuclei exhibited a different pattern of tritium incorporation from that observed in G1 or G2 nuclei (×100 oil immersion). Because nuclear membranes were not always visible on photomicrographs, the nucleus is outlined in red. Note the visualization of replicon clusters in G1/S transitional nuclei, while a G1 nucleus exhibits only UDS. The number of UDS grains per nucleus was quantitated, providing a measure of the number of sites of repair synthesis per nucleus. In comparing wild-type and (+/+) mutant MEFs, differences were observed in numbers of UDS grains per nucleus. UDS experiments were terminated 3 h after UV radiation (compare UDS results to the 4-h time point shown in Fig. 2). (C to E) Compiled data from several experiments such as those shown in panel B. Values obtained for wild-type cells were defined as 100%. (C) Compiled data for UDS after UV irradiation. The following numbers of nuclei were counted: wild-type, 652; _p53_−/−, 88; _p21_−/−, 129; _gadd45_−/−, 177; _gadd45/p21_-null, 157. The values shown are means ± standard deviations. (D) Compiled data, control MEF lines, UDS after UV irradiation. The following numbers of nuclei were counted: wild-type, 51; _rb_−/−, 22; _p16_−/−, 20. The values shown are means ± standard deviations. (E) Compiled data for UDS after cisplatin treatment. The following numbers of nuclei were counted: wild-type, 116; _p53_−/−, 93; gadd45/p21, 81. The values shown are means ± standard deviations. (F) PCNA immunostaining (Triton resistant) in wild-type and _gadd45_−/− MEFs 1.5 h after UV radiation (top) or 3 h after UV radiation (bottom). While all wild-type nuclei were clearly visible, _gadd45_−/− nuclei were much less visible; their positions are marked by arrows. The PCNA staining defect was most evident at 1.5 h and recovered to near normal after 6 h (not shown).
FIG. 1
DNA repair deficiency in cells lacking p53 or p53 effector genes, gadd45 and p21, measured by UDS. (A) Illustration of UDS technique. A low-magnification (×20) field of cells irradiated with 20 J of UV radiation m−2 is shown. Nuclei were made visible by the incorporation of [3H]thymidine during either replicative or repair DNA synthesis. S-phase nuclei appear black upon the photographic emulsion, while non-S-phase cells exhibit UDS. The _p21_−/− and _gadd45/p21_-null lines did exhibit a higher S-phase fraction (50%) compared to wild-type or _gadd45_−/− lines (15 to 20%). (B) Distinctions between replicative DNA synthesis and repair synthesis (UDS). S-phase or G1/S transitional nuclei exhibited a different pattern of tritium incorporation from that observed in G1 or G2 nuclei (×100 oil immersion). Because nuclear membranes were not always visible on photomicrographs, the nucleus is outlined in red. Note the visualization of replicon clusters in G1/S transitional nuclei, while a G1 nucleus exhibits only UDS. The number of UDS grains per nucleus was quantitated, providing a measure of the number of sites of repair synthesis per nucleus. In comparing wild-type and (+/+) mutant MEFs, differences were observed in numbers of UDS grains per nucleus. UDS experiments were terminated 3 h after UV radiation (compare UDS results to the 4-h time point shown in Fig. 2). (C to E) Compiled data from several experiments such as those shown in panel B. Values obtained for wild-type cells were defined as 100%. (C) Compiled data for UDS after UV irradiation. The following numbers of nuclei were counted: wild-type, 652; _p53_−/−, 88; _p21_−/−, 129; _gadd45_−/−, 177; _gadd45/p21_-null, 157. The values shown are means ± standard deviations. (D) Compiled data, control MEF lines, UDS after UV irradiation. The following numbers of nuclei were counted: wild-type, 51; _rb_−/−, 22; _p16_−/−, 20. The values shown are means ± standard deviations. (E) Compiled data for UDS after cisplatin treatment. The following numbers of nuclei were counted: wild-type, 116; _p53_−/−, 93; gadd45/p21, 81. The values shown are means ± standard deviations. (F) PCNA immunostaining (Triton resistant) in wild-type and _gadd45_−/− MEFs 1.5 h after UV radiation (top) or 3 h after UV radiation (bottom). While all wild-type nuclei were clearly visible, _gadd45_−/− nuclei were much less visible; their positions are marked by arrows. The PCNA staining defect was most evident at 1.5 h and recovered to near normal after 6 h (not shown).
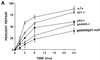
FIG. 2
Deficient UV photoproduct repair in genomic DNA isolated from _p53_- or _gadd45_-null MEF lines. (A) Kinetics of 6-4 pp removal in MEFs determined by immunoassays following 10 J of UV irradiation m−2. Data were from two independent experiments conducted in triplicate. The values shown are means ± standard deviations. Note that 100% repair was defined relative to unirradiated cells. The bulk of the damage is repaired in wild-type cells within the first 3 to 4 h (compare the 4-h time point with the UDS data in Fig. 1). (B) Strand-specific repair of CPDs within the dhfr gene measured by TEV assays following 10 J of UV irradiation m−2. Repair of the NTS (red) was markedly reduced in _gadd45_−/− or _gadd45/p21_-null MEFs. Each of the MEF lines exhibited near normal TCR repair of the TS (blue).
FIG. 2
Deficient UV photoproduct repair in genomic DNA isolated from _p53_- or _gadd45_-null MEF lines. (A) Kinetics of 6-4 pp removal in MEFs determined by immunoassays following 10 J of UV irradiation m−2. Data were from two independent experiments conducted in triplicate. The values shown are means ± standard deviations. Note that 100% repair was defined relative to unirradiated cells. The bulk of the damage is repaired in wild-type cells within the first 3 to 4 h (compare the 4-h time point with the UDS data in Fig. 1). (B) Strand-specific repair of CPDs within the dhfr gene measured by TEV assays following 10 J of UV irradiation m−2. Repair of the NTS (red) was markedly reduced in _gadd45_−/− or _gadd45/p21_-null MEFs. Each of the MEF lines exhibited near normal TCR repair of the TS (blue).
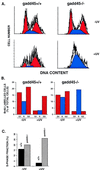
FIG. 3
Pronounced S-phase delay following UV-damage in _gadd45_−/− MEFs. (A) Flow cytometric profiles shown by PI staining. In the absence of UV irradiation, cell cycle profiles were similar (50 to 60% G1, 10 to 25% S, 20 to 35% G2/M). G1 and G2 peaks are in shown in red; the S-phase fraction is shown in blue. After UV irradiation, _gadd45_−/− MEFs exhibited a marked S-phase delay. The results shown correspond to 24 h after 10 J of UV irradiation m−2. Only BrdU-positive cells that were actively cycling are shown (10). The gating of BrdU-positive cells was designed to exclude cells arrested in the first G1, which as cited in other studies, was not affected by the presence or absence of Gadd45. The technique further distinquishes the p53/gadd45-mediated response to UV radiation from p53/p21-mediated G1 arrest, as discussed in the text. (B) Summary of cell cycle profiles in MEFs in the absence or presence of UV radiation. The results shown in panel A are summarized by bar graphs. Again, _gadd45_−/− cells are delayed in S-phase progression. Results shown correspond to 24 h after 10 J of UV irradiation m−2. Only BrdU-positive cells that were actively cycling are shown (10). (C) Determination of S-phase fractions 15 h after UV irradiation by [3H]thymidine pulse-labelling. UV-irradiated _gadd45_−/− MEFs exhibit a pronounced S-phase fraction 15 h after UV damage. The results were obtained from two or more independent experiments.
FIG. 4
(A) Gadd45 affects in vitro NER-chromatin assembly in experiments with mouse cell extracts. WCE and NUC extracts prepared from wild-type (+/+) or _gadd45_−/− (−/−) mouse lymphoblasts were incubated with UV-damaged plasmid template (left panel). While little form I DNA was recovered from the reactions, evidenced by EtBr staining (total DNA), a greater fraction of repaired (radiolabeled) DNA was recovered as form I. Gadd45 affects the recovery of form I (repaired, radiolabeled) DNA, which is ordered into nucleosome ladders, evidenced by MNase digestion (shown in panel B). Gadd45 was either endogenous to wild-type extracts, or recombinant Gadd45 (rGadd45) was added to _gadd45_−/− extracts in the amounts indicated (micrograms). (Right panel) Plasmid templates lacking UV damage exhibited low levels of 32P labelling. Lanes show approximate 32P incorporation as follows: 1 to 4, 150 fmol; 6, 190 fmol; 5 and 7, 11 fmol. Form II DNA was predominantly nicked, while form I DNA was predominantly supercoiled. (B) MNase digestion of plasmids recovered from in vitro NER-chromatin assembly assays, corresponding to lanes 2, 4, and 5 in panel A. In the absence of Gadd45 (lane 2), the predominantly form II plasmid DNA did not exhibit nucleosome ladders after MNase digestion. In the presence of Gadd45 (lane 4), laddering was observed, indicative of chromatin assembly. Undamaged plasmids were not assembled into chromatin (lane 5).
Similar articles
- Human cells deficient in p53 regulated p21(waf1/cip1) expression exhibit normal nucleotide excision repair of UV-induced DNA damage.
Wani MA, Wani G, Yao J, Zhu Q, Wani AA. Wani MA, et al. Carcinogenesis. 2002 Mar;23(3):403-10. doi: 10.1093/carcin/23.3.403. Carcinogenesis. 2002. PMID: 11895854 - Loss of p21WAF1/Cip1 in Gadd45-deficient keratinocytes restores DNA repair capacity.
Maeda T, Espino RA, Chomey EG, Luong L, Bano A, Meakins D, Tron VA. Maeda T, et al. Carcinogenesis. 2005 Oct;26(10):1804-10. doi: 10.1093/carcin/bgi140. Epub 2005 May 25. Carcinogenesis. 2005. PMID: 15917306 - Abrogation of p53 function affects gadd gene responses to DNA base-damaging agents and starvation.
Zhan Q, Fan S, Smith ML, Bae I, Yu K, Alamo I Jr, O'Connor PM, Fornace AJ Jr. Zhan Q, et al. DNA Cell Biol. 1996 Oct;15(10):805-15. doi: 10.1089/dna.1996.15.805. DNA Cell Biol. 1996. PMID: 8892753 - Gadd45 in the response of hematopoietic cells to genotoxic stress.
Liebermann DA, Hoffman B. Liebermann DA, et al. Blood Cells Mol Dis. 2007 Nov-Dec;39(3):329-35. doi: 10.1016/j.bcmd.2007.06.006. Epub 2007 Jul 30. Blood Cells Mol Dis. 2007. PMID: 17659913 Free PMC article. Review. - Cell cycle checkpoints and DNA repair preserve the stability of the human genome.
Kaufmann WK. Kaufmann WK. Cancer Metastasis Rev. 1995 Mar;14(1):31-41. doi: 10.1007/BF00690209. Cancer Metastasis Rev. 1995. PMID: 7606819 Review.
Cited by
- Gadd45 in DNA Demethylation and DNA Repair.
Chandramouly G. Chandramouly G. Adv Exp Med Biol. 2022;1360:55-67. doi: 10.1007/978-3-030-94804-7_4. Adv Exp Med Biol. 2022. PMID: 35505162 - p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene.
Adimoolam S, Ford JM. Adimoolam S, et al. Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12985-90. doi: 10.1073/pnas.202485699. Epub 2002 Sep 19. Proc Natl Acad Sci U S A. 2002. PMID: 12242345 Free PMC article. - Significance analysis of microarrays applied to the ionizing radiation response.
Tusher VG, Tibshirani R, Chu G. Tusher VG, et al. Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):5116-21. doi: 10.1073/pnas.091062498. Epub 2001 Apr 17. Proc Natl Acad Sci U S A. 2001. PMID: 11309499 Free PMC article. - Gadd45 in stress signaling.
Liebermann DA, Hoffman B. Liebermann DA, et al. J Mol Signal. 2008 Sep 12;3:15. doi: 10.1186/1750-2187-3-15. J Mol Signal. 2008. PMID: 18789159 Free PMC article. - TAp63γ enhances nucleotide excision repair through transcriptional regulation of DNA repair genes.
Liu J, Lin M, Zhang C, Wang D, Feng Z, Hu W. Liu J, et al. DNA Repair (Amst). 2012 Feb 1;11(2):167-76. doi: 10.1016/j.dnarep.2011.10.016. Epub 2011 Nov 6. DNA Repair (Amst). 2012. PMID: 22056305 Free PMC article.
References
- Aboussekhra A, Biggerstaff M, Shivji M, Vilpo J, Moncollin V, Podust V, Protic M, Hubscher U, Egly J-M, Wood R D. Mammalian DNA excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
- Aboussekhra A, Wood R D. Detection of nucleotide excision repair incisions in human fibroblasts by immunostaining for PCNA. Exp Cell Res. 1995;221:326–332. - PubMed
- Baxter B K, Smerdon M J. Nucleosome unfolding during DNA repair in normal and xeroderma pigmentosum (group C) human cells. J Biol Chem. 1998;273:17517–17524. - PubMed
- Brown J M, Wouters B G. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–1399. - PubMed
- Burk P G, Lutzner M A, Clarke D D, Robbins J H. Ultraviolet-stimulated thymidine incorporation in xeroderma pigmentosum lymphoblasts. J Lab Clin Med. 1971;77:759–767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous