A decamer duplication in the 3' region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred - PubMed (original) (raw)
A decamer duplication in the 3' region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred
R Vidal et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Familial Danish dementia (FDD), also known as heredopathia ophthalmo-oto-encephalica, is an autosomal dominant disorder characterized by cataracts, deafness, progressive ataxia, and dementia. Neuropathological findings include severe widespread cerebral amyloid angiopathy, hippocampal plaques, and neurofibrillary tangles, similar to Alzheimer's disease. N-terminal sequence analysis of isolated leptomeningeal amyloid fibrils revealed homology to ABri, the peptide originated by a point mutation at the stop codon of gene BRI in familial British dementia. Molecular genetic analysis of the BRI gene in the Danish kindred showed a different defect, namely the presence of a 10-nt duplication (795-796insTTTAATTTGT) between codons 265 and 266, one codon before the normal stop codon 267. The decamer duplication mutation produces a frame-shift in the BRI sequence generating a larger-than-normal precursor protein, of which the amyloid subunit (designated ADan) comprises the last 34 C-terminal amino acids. This de novo-created amyloidogenic peptide, associated with a genetic defect in the Danish kindred, stresses the importance of amyloid formation as a causative factor in neurodegeneration and dementia.
Figures
Figure 1
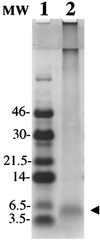
Isolation of ADan. Tris⋅Tricine SDS/PAGE (16%) of the amyloid protein isolated from leptomeningeal vessels stained with Coomassie blue. The 4-kDa band (line 2, arrowhead) was subjected to Edman N-terminal amino acid sequence analysis after being transferred onto an Immobilon-P membrane. Line 1: Low molecular-mass markers (Amersham) in kDa.
Figure 2
Analysis of PCR-amplified genomic DNA in the Danish kindred. (A) Polyacrylamide gel showing the restriction enzyme analysis performed as described on ref. . Genomic DNA was amplified with oligonucleotides F and R and analyzed with _Xba_I from a normal control (lane 1), a patient with FDD (lane 2), and a patient with FBD (lane 3). The arrowhead indicates undigested DNA (191 bp), and the arrows indicate digested fragments of 116 and 75 bp. (B) Metaphor agarose gel (4%) of PCR-amplified fragments by using primers F and R2. Lane 1: 123 molecular-mass marker. Lane 2: PCR product of amplification of the BRI gene in a patient with FDD. Lane 3: PCR amplification of a normal control. Amplification in the same conditions by using genomic DNA from patients with FBD gave identical results as seen in normal controls. The extra band observed in the Danish kindred (151 bp) is indicated by an arrow.
Figure 3
Nucleotide and amino acid sequence of ADan. Nucleotide and translated amino acid sequence of the wild-type BRI gene between nucleotides 730 to 824 and the corresponding sequence in amyloid ADan. A gap in the wild-type BRI sequence was added to show the insertion of the 10 nucleotides that are duplicated (underlined) in FDD patients. Nucleotides are numbered in the 5′ to 3′ orientation. Termination codons are noted in bold. Amino acid sequences are in one-letter code.
Figure 4
MS and Western blot analysis of leptomeningeal ADan. (A) MS analysis of the leptomeningeal amyloid isolated from the Danish case. * indicates formylated species (+28 units of mass), and @ indicates species with pyroglutamate at the amino terminus (−18 units of mass). The sequence of the full-length amyloid (34 amino acids) is indicated in one-letter code. (B) Western blotting analysis of purified amyloid fibrils from leptomeningeal vessels of cases with FDD (lanes 1 and 4) and FBD (lanes 2 and 3) reacted against anti-ADan antibody (5282) (lanes 1 and 2) and anti-ABri antibody (338) (lanes 3 and 4). Molecular masses are in kDa.
Figure 5
Immunohistochemical analysis. Parenchymal vessels of temporal sections (A) immunostained with Ab 5282 (×40) and (B) observed under polarized light after Congo red staining (×40). Examples of amyloid angiopathy with perivascular deposits in the subiculum (C) and granular cell layer of the cerebellar cortex (D) (Ab 5282; ×200). All regions of the hippocampus contain numerous Ab 5282-positive plaques (E) giving a good outline of the anatomy of the hippocampus (Ab 5282; ×4). Cotton wool-like plaques are present in the CA4 subregion of the hippocampus (F), whereas confluent looser deposits are characteristic in other subregions including CA1 (G) (Ab 5282; ×125). Cerebellum showed positivity in leptomeninges, leptomeningeal, and parenchymal blood vessels (H), with a perivascular plaque around the blood vessel in the granular cell layer (Ab 5282; ×30). Paraffin sections, hematoxylin counter stain.
Figure 6
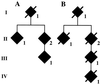
Analysis of the 10-nt duplication in the BRI precursor protein gene in the Danish kindred. Members of the Danish family pedigree are shown in A and B. Roman numbers indicate different generations. Only affected members (filled symbols) are indicated. Genomic DNA was isolated from blood samples of cases AII1, AII2, and AIII1, and from formalin tissue of samples BII2 and BIII1 and from brain tissue of case IV1. In all cases, the 10-nt duplication was determined after cloning and sequencing of the amplified DNA.
Figure 7
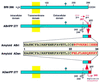
Schematic representation of the amyloid precursor proteins and amyloid peptides in patients affected with FBD and FDD. The BRI gene codifies a putative single transmembrane-type II precursor protein of 266 amino acids. Different genetic defects in the BRI gene, namely a point mutation in FBD (Stop-267→Arg) and a 10-nt duplication (795–796ins TTTAATTTGT) in FDD, originate the two precursor molecules ABriPP and ADanPP, respectively. Although originated by different genetic defects in the BRI gene, both mutated precursors have 277 amino acids. The scissors indicate the cleavage site of the precursor molecules between codons 243 (Arg) and 244 (Glu) that releases the ABri and ADan peptides. Both amyloids have identical N-terminal amino acid sequence (first 22 amino acids) and a completely different C-terminal sequence (last 12 amino acids). The diamond indicates the single glycosylation site at position 170.
Similar articles
- A stop-codon mutation in the BRI gene associated with familial British dementia.
Vidal R, Frangione B, Rostagno A, Mead S, Révész T, Plant G, Ghiso J. Vidal R, et al. Nature. 1999 Jun 24;399(6738):776-81. doi: 10.1038/21637. Nature. 1999. PMID: 10391242 - Properties of neurotoxic peptides related to the Bri gene.
El-Agnaf O, Gibson G, Lee M, Wright A, Austen BM. El-Agnaf O, et al. Protein Pept Lett. 2004 Jun;11(3):207-12. doi: 10.2174/0929866043407156. Protein Pept Lett. 2004. PMID: 15182222 - Cerebral amyloid angiopathy and parenchymal amyloid deposition in transgenic mice expressing the Danish mutant form of human BRI2.
Vidal R, Barbeito AG, Miravalle L, Ghetti B. Vidal R, et al. Brain Pathol. 2009 Jan;19(1):58-68. doi: 10.1111/j.1750-3639.2008.00164.x. Epub 2008 Apr 10. Brain Pathol. 2009. PMID: 18410407 Free PMC article. - Chromosome 13 dementia syndromes as models of neurodegeneration.
Ghiso J, Révész T, Holton J, Rostagno A, Lashley T, Houlden H, Gibb G, Anderton B, Bek T, Bojsen-Møller M, Wood N, Vidal R, Braendgaard H, Plant G, Frangione B. Ghiso J, et al. Amyloid. 2001 Dec;8(4):277-84. doi: 10.3109/13506120108993826. Amyloid. 2001. PMID: 11791622 Review. - Chromosome 13 dementias.
Rostagno A, Tomidokoro Y, Lashley T, Ng D, Plant G, Holton J, Frangione B, Revesz T, Ghiso J. Rostagno A, et al. Cell Mol Life Sci. 2005 Aug;62(16):1814-25. doi: 10.1007/s00018-005-5092-5. Cell Mol Life Sci. 2005. PMID: 15968464 Free PMC article. Review.
Cited by
- Translation readthrough mitigation.
Arribere JA, Cenik ES, Jain N, Hess GT, Lee CH, Bassik MC, Fire AZ. Arribere JA, et al. Nature. 2016 Jun 30;534(7609):719-23. doi: 10.1038/nature18308. Epub 2016 Jun 1. Nature. 2016. PMID: 27281202 Free PMC article. - The Bri2 and Bri3 BRICHOS Domains Interact Differently with Aβ42 and Alzheimer Amyloid Plaques.
Dolfe L, Tambaro S, Tigro H, Del Campo M, Hoozemans JJM, Wiehager B, Graff C, Winblad B, Ankarcrona M, Kaldmäe M, Teunissen CE, Rönnbäck A, Johansson J, Presto J. Dolfe L, et al. J Alzheimers Dis Rep. 2018 Feb 16;2(1):27-39. doi: 10.3233/ADR-170051. J Alzheimers Dis Rep. 2018. PMID: 30480246 Free PMC article. - The Amyloid-Tau-Neuroinflammation Axis in the Context of Cerebral Amyloid Angiopathy.
Cisternas P, Taylor X, Lasagna-Reeves CA. Cisternas P, et al. Int J Mol Sci. 2019 Dec 14;20(24):6319. doi: 10.3390/ijms20246319. Int J Mol Sci. 2019. PMID: 31847365 Free PMC article. Review. - Expression of BRI2 mRNA and protein in normal human brain and familial British dementia: its relevance to the pathogenesis of disease.
Lashley T, Revesz T, Plant G, Bandopadhyay R, Lees AJ, Frangione B, Wood NW, de Silva R, Ghiso J, Rostagno A, Holton JL. Lashley T, et al. Neuropathol Appl Neurobiol. 2008 Oct;34(5):492-505. doi: 10.1111/j.1365-2990.2008.00935.x. Epub 2008 Feb 13. Neuropathol Appl Neurobiol. 2008. PMID: 18282158 Free PMC article. - Recombinant Bri3 BRICHOS domain is a molecular chaperone with effect against amyloid formation and non-fibrillar protein aggregation.
Poska H, Leppert A, Tigro H, Zhong X, Kaldmäe M, Nilsson HE, Hebert H, Chen G, Johansson J. Poska H, et al. Sci Rep. 2020 Jun 17;10(1):9817. doi: 10.1038/s41598-020-66718-y. Sci Rep. 2020. PMID: 32555390 Free PMC article.
References
- Strömgrem E, Dalby A, Dalby M, Ranheim B. Acta Neurol Scand. 1970;46,(Suppl. 43):97–98. - PubMed
- Strömgrem E. In: Handbook of Clinical Neurology. Vinken P J, Bruyn G W, editors. Amsterdam: Elsevier; 1981. pp. 150–152.
- Plant G, Esiri M M. In: The Neuropathology of Dementia. Esiri M M, Morris J H, editors. Cambridge, U.K.: Cambridge Univ. Press; 1997. pp. 260–276.
- Vidal R, Frangione B, Rostagno A, Mead S, Révész T, Plant G, Ghiso J. Nature (London) 1999;399:776–781. - PubMed
- Deleersnijder W, Hong G, Cortvrindt R, Poirier C, Tylzanowski P, Pittois K, Van Marck E, Merregaert J. J Biol Chem. 1996;271:19475–19482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG10953/AG/NIA NIH HHS/United States
- R01 AG008721/AG/NIA NIH HHS/United States
- AG05891/AG/NIA NIH HHS/United States
- AG08721/AG/NIA NIH HHS/United States
- R37 AG005891/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous