MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M - PubMed (original) (raw)
MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M
J Zhu et al. Mol Cell Biol. 2000 Aug.
Abstract
p53, a tumor suppressor, inhibits cell proliferation by inducing cellular genes involved in the regulation of the cell cycle. MCG10, a novel cellular p53 target gene, was identified in a cDNA subtraction assay with mRNA isolated from a p53-producing cell line. MCG10 can be induced by wild-type but not mutant p53 and by DNA damage via two potential p53-responsive elements in the promoter of the MCG10 gene. The MCG10 gene contains 10 exons and is located at chromosome 3p21, a region highly susceptible to aberrant chromosomal rearrangements and deletions in human neoplasia. The MCG10 gene locus encodes at least two alternatively spliced transcripts, MCG10 and MCG10as. The MCG10 and MCG10as proteins contain two domains homologous to the heterogeneous nuclear ribonucleoprotein K homology (KH) domain. By generating cell lines that inducibly express either wild-type or mutated forms of MCG10 and MCG10as, we found that MCG10 and MCG10as can suppress cell proliferation by inducing apoptosis and cell cycle arrest in G(2)-M. In addition, we found that MCG10 and MCG10as, through their KH domains, can bind poly(C) and that their RNA-binding activity is necessary for inducing apoptosis and cell cycle arrest. Furthermore, we found that the level of the poly(C) binding MCG10 protein is increased in cells treated with the DNA-damaging agent camptothecin in a p53-dependent manner. These results suggest that the MCG10 RNA-binding protein is a potential mediator of p53 tumor suppression.
Figures
FIG. 1
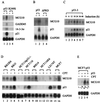
Upregulation of MCG10 by p53. (A) Wild-type p53 but not mutant p53 induces MCG10. Northern blots were prepared using 10 μg of total RNA isolated from p53-3 or p53(R249S) cells that were uninduced (−) or induced (+) to express wild-type p53 or mutant p53(R249S), respectively. The blots were probed with cDNAs derived from the MCG10, 14-3-3ς, GADD45, p21 and GAPDH genes. (B) The apoptosis-deficient deletion mutant p53(ΔPRD) is incapable of inducing MCG10. A Northern blot was prepared using 10 μg of total RNA isolated from p53-3 or p53(ΔPRD)-5 cells that were uninduced (−) or induced (+) to express wild-type p53 or mutant p53(ΔPRD), respectively. The blot was probed with MCG10 cDNA and then reprobed with both p21 and GAPDH cDNAs. (C) Kinetics of p53 induction of MCG10. A Northern blot was prepared using 10 μg of total RNA isolated from p53-3 cells that were induced for 0, 6, 12, 18, 24, 36, or 48 h. The blot was probed with MCG10 cDNA and then reprobed with both p21 and GAPDH cDNAs. (D) MCG10 is induced by DNA damage in cells that contain an endogenous wild-type p53 gene but not in cells that are functionally p53-null. Northern blots were prepared using 10 μg of total RNA isolated from seven individual cell lines (see text for details) as indicated above the figure, which were untreated (−) or treated (+) with 300 nM camptothecin for 24 h. The blots were probed with cDNAs derived from MCG10, p21, and GAPDH. (E) Exogenous inducible p53 cooperates with endogenous wild-type p53 in MCF7 cells to induce MCG10. A Northern blot was prepared using 10 μg of total RNA isolated from MCF7-p53 cells that were untreated (lane 1), treated with 300 nM camptothecin (CPT) to induce endogenous wild-type p53 (lane 2), induced to express exogenous p53 and treated with 300 nM camptothecin to induce endogenous wild-type p53 (lane 3), or induced to express exogenous p53 (lane 4). The blot was probed with cDNAs derived from MCG10 and GAPDH.
FIG. 2
Identification of two p53-responsive elements in the MCG10 gene. (A) Schematic representation of the MCG10 genomic DNA structure. Exons are shown as numbered boxes, and introns are shown as lines. The locations of two potential p53-responsive elements in the promoter of the MCG10 gene are indicated. Bold uppercase letters represent nucleotides predicted to be critical for the consensus p53-responsive element. Lowercase letters represent mismatches. The transcript for MCG10 is drawn above the gene structure, and the transcript for MCG10as is shown below. Exon 4b is not present in the MCG10as transcript. (B) Two of the three potential p53-binding sites but not their mutated forms in the MCG10 gene are responsive to wild-type p53 in vivo. p53RE-1, m-p53RE-1, p53RE-2, m-p53RE-2, or p53RE-3 (2 μg) was cotransfected into H1299 cells with 1 μg of pcDNA3 control vector or a vector that expresses wild-type p53 or mutant p53(R175H). The fold increase in relative luciferase activity is the luciferase activity induced by p53 divided by that induced by pcDNA3. Error bars represent the standard deviations from at least three experiments. (C) p53 binds specifically to both p53RE-1 and p53RE-2 in vitro. The 28- and 36-bp oligonucleotide fragments containing p53RE-1 and p53RE-2, respectively, in the MCG10 gene were labeled with [α-32P]dCTP. The labeled probe DNA (5 ng) was added to a mixture containing 20 ng of p53 protein. The p53-DNA complex was resolved in a 4% polyacrylamide gel. For competition assays, 5- or 20-fold excess unlabeled 28-bp probe DNA (lanes 3 and 4), 36-bp unlabeled probe DNA (lanes 9 and 10), or RGC (lanes 5 and 6 and 11 and 12) was added to the reactions.
FIG. 2
Identification of two p53-responsive elements in the MCG10 gene. (A) Schematic representation of the MCG10 genomic DNA structure. Exons are shown as numbered boxes, and introns are shown as lines. The locations of two potential p53-responsive elements in the promoter of the MCG10 gene are indicated. Bold uppercase letters represent nucleotides predicted to be critical for the consensus p53-responsive element. Lowercase letters represent mismatches. The transcript for MCG10 is drawn above the gene structure, and the transcript for MCG10as is shown below. Exon 4b is not present in the MCG10as transcript. (B) Two of the three potential p53-binding sites but not their mutated forms in the MCG10 gene are responsive to wild-type p53 in vivo. p53RE-1, m-p53RE-1, p53RE-2, m-p53RE-2, or p53RE-3 (2 μg) was cotransfected into H1299 cells with 1 μg of pcDNA3 control vector or a vector that expresses wild-type p53 or mutant p53(R175H). The fold increase in relative luciferase activity is the luciferase activity induced by p53 divided by that induced by pcDNA3. Error bars represent the standard deviations from at least three experiments. (C) p53 binds specifically to both p53RE-1 and p53RE-2 in vitro. The 28- and 36-bp oligonucleotide fragments containing p53RE-1 and p53RE-2, respectively, in the MCG10 gene were labeled with [α-32P]dCTP. The labeled probe DNA (5 ng) was added to a mixture containing 20 ng of p53 protein. The p53-DNA complex was resolved in a 4% polyacrylamide gel. For competition assays, 5- or 20-fold excess unlabeled 28-bp probe DNA (lanes 3 and 4), 36-bp unlabeled probe DNA (lanes 9 and 10), or RGC (lanes 5 and 6 and 11 and 12) was added to the reactions.
FIG. 3
(A and B) Deduced amino acid sequences of MCG10 and MCG10as. The N-terminal KH domain (KH1) and C-terminal KH domain (KH2) in MCG10 and MCG10as are boxed. The bold italic letters represent a 55-amino-acid insertion in the N-terminal KH domain of MCG10. Three proline-rich domains (PRD) are underlined. The nuclear export signal (NES) and nuclear localization signal (NLS) are marked by dashes. (C) Schematic representations of MCG10 and MCG10as protein structures. The locations of specific features are indicated by the amino acid number. (D) Sequence alignment of eight KH domains from hnRNP K, FMR1, MCG10, and MCG10as. Numbers on the right indicate positions of the ending amino acids in the KH domain. Highly conserved positions are highlighted in colors. The GXXG motif is shown below the alignment. The critical isoleucine residue for FMR1 KH2 that is mutated in fragile X syndrome is indicated (∗).
FIG. 3
(A and B) Deduced amino acid sequences of MCG10 and MCG10as. The N-terminal KH domain (KH1) and C-terminal KH domain (KH2) in MCG10 and MCG10as are boxed. The bold italic letters represent a 55-amino-acid insertion in the N-terminal KH domain of MCG10. Three proline-rich domains (PRD) are underlined. The nuclear export signal (NES) and nuclear localization signal (NLS) are marked by dashes. (C) Schematic representations of MCG10 and MCG10as protein structures. The locations of specific features are indicated by the amino acid number. (D) Sequence alignment of eight KH domains from hnRNP K, FMR1, MCG10, and MCG10as. Numbers on the right indicate positions of the ending amino acids in the KH domain. Highly conserved positions are highlighted in colors. The GXXG motif is shown below the alignment. The critical isoleucine residue for FMR1 KH2 that is mutated in fragile X syndrome is indicated (∗).
FIG. 4
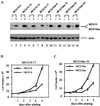
MCG10 and MCG10as are capable of suppressing cell proliferation. (A) Levels of MCG10, MCG10as, and actin were assayed by Western blot analysis in cell lines that inducibly express MCG10 or MCG10as. Cell extracts were prepared from uninduced cells (−) or cells induced (+) to express MCG10 or MCG10as. The blot was probed with affinity-purified anti-MCG10 polyclonal antibody (upper panel) and then reprobed with antiactin polyclonal antibody (lower panel). (B and C) Growth rates of MCG10-17 and MCG10as-10 cells in the presence (◊) or absence (□) of MCG10 or MCG10as, respectively, were measured as described in Materials and Methods. Error bars represent the standard deviations from at least three experiments.
FIG. 5
MCG10 is capable of inducing cell cycle arrest in G2-M and apoptosis. DNA content was quantitated by propidium iodide staining of fixed cells that were uninduced (−MCG10) or induced (+MCG10) to express MCG10 for 2 days (A and B), 4 days (E and F), and 6 days (I and J). Apoptotic cells were quantitated by propidium iodide-annexin V staining of cells that were uninduced (−MCG10) or induced (+MCG10) to express MCG10 for 2 days (C and D), 4 days (G and H), and 6 days (K and L).
FIG. 6
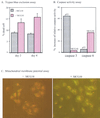
MCG10 activates caspase 6 and induces apoptosis through the mitochondrial pathway. (A) The percentage of dead cells induced by MCG10 was quantified by trypan blue dye exclusion. Cells were seeded in the presence (+) or absence (−) of MCG10 for 2 or 4 days. Both unstained and trypan blue-stained cells were counted using a hemocytometer. Error bars represent the standard deviations from at least three experiments. (B) Caspase 6 is activated by MCG10. p53-3 or MCG10-17 cells were uninduced or induced to express p53 or MCG10 for 3 days. Cells were then collected and assayed for the activity of caspases 3 and 6 as described in Materials and Methods. (C) The mitochondrial membrane potentials were altered in cells induced to express MCG10. MCG10-17 cells were uninduced (−MCG10) or induced to express MCG10 (+MCG10) for 3 days, stained with Mitosensor, and analyzed by fluorescence microscopy.
FIG. 7
MCG10as is capable of inducing both cell cycle arrest in G2-M and apoptosis. DNA content was quantitated by propidium iodide staining of fixed cells that were uninduced (−MCG10as) or induced (+MCG10as) to express MCG10as for 2 days (A and B), 4 days (E and F), and 6 days (I and J). Apoptotic cells were quantitated by propidium iodide-annexin V staining of cells that were uninduced (−MCG10as) or induced (+MCG10as) to express MCG10as for 2 days (C and D), 4 days (G and H), and 6 days (K and L).
FIG. 8
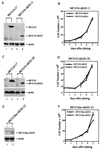
Both KH domains in MCG10 and MCG10as are necessary for inducing cell cycle arrest and apoptosis. (A) Levels of MCG10 and actin in MCG10-17 and MCG10-ΔKH1-5 cell lines were assayed by Western blot analysis. Cell extracts were prepared from uninduced cells (−) or cells induced (+) to express MCG10 or MCG10-ΔKH1. The blot was probed with affinity-purified anti-MCG10 polyclonal antibody (upper panel) and then reprobed with antiactin polyclonal antibody (lower panel). (B) Growth rates of MCG10-ΔKH1-5 cells in the presence (◊) and absence (□) of MCG10-ΔKH1 were measured as described in Materials and Methods. (C) Levels of MCG10 and actin in MCG10-17 and MCG10-ΔKH2-20 cell lines were assayed by Western blot analysis as described for panel A. (D) Growth rates of MCG10-ΔKH2-20 cells in the presence (◊) and absence (□) of MCG10-ΔKH2. (E) Levels of MCG10as-ΔKH2 and actin in the MCG10as-ΔKH2-23 cell line were assayed by Western blot analysis as described for panel A. (F) Growth rates of MCG10as-ΔKH2-23 cells in the presence (◊) and absence (□) of MCG10as-ΔKH2. Error bars represent the standard deviations from at least three experiments.
FIG. 9
KH domain in MCG10 and MCG10as is capable of and necessary for binding poly(C). (A) MCG10 and MCG10as can bind to poly(C) but not to poly(U), poly(G), or poly(A). Total cell extracts run in lanes 1 and 2 were prepared from MCG10-17 and MCG10as-10 cells that were uninduced (−) and induced (+) to express MCG10 (upper panel) or MCG10as (lower panel). Cytoplasmic extracts (C) and nuclear extracts (N) were prepared from uninduced cells (−) or cells induced (+) to express MCG10 or MCG10as and mixed with poly(C)-, poly(U)-, poly(G)-, or poly(A)-agarose beads. Proteins bound to the beads were isolated and assayed by Western blot analysis using anti-MCG10 antibody. (B) The KH1 domain in MCG10 is necessary for binding poly(C). Cytoplasmic extracts were prepared from cells induced to express MCG10-ΔKH1, and the RNA-binding assay was performed as described for panel A. (C and D) The KH2 domain in MCG10 and MCG10as is necessary for binding poly(C). Cytoplasmic extracts were prepared from cells induced to express MCG10-ΔKH2 or MCG10as-ΔKH2, and the RNA-binding assay was performed. (E) A point mutation (Ile230Asp) in the KH2 domain abrogates the ability of MCG10as to bind to poly(C). Cytoplasmic extracts were prepared from cells induced to express MCG10as-KH2−, and the RNA-binding assay was performed.
FIG. 10
Level of the poly(C)-binding MCG10 protein is increased in cells treated with DNA-damaging agent camptothecin in a p53-dependent manner. Cytoplasmic extracts were prepared from RKO, RKOE6, HCT116, and HCT116E6 cells that were untreated (−) or treated (+) with camptothecin (CPT). The RNA-binding assay was performed as described in the legend to Fig. 9A.
Similar articles
- Dissociation between cell cycle arrest and apoptosis can occur in Li-Fraumeni cells heterozygous for p53 gene mutations.
Delia D, Goi K, Mizutani S, Yamada T, Aiello A, Fontanella E, Lamorte G, Iwata S, Ishioka C, Krajewski S, Reed JC, Pierotti MA. Delia D, et al. Oncogene. 1997 May 8;14(18):2137-47. doi: 10.1038/sj.onc.1201050. Oncogene. 1997. PMID: 9174049 - Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains.
Dejgaard K, Leffers H. Dejgaard K, et al. Eur J Biochem. 1996 Oct 15;241(2):425-31. doi: 10.1111/j.1432-1033.1996.00425.x. Eur J Biochem. 1996. PMID: 8917439 - 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis.
Oh JJ, Razfar A, Delgado I, Reed RA, Malkina A, Boctor B, Slamon DJ. Oh JJ, et al. Cancer Res. 2006 Apr 1;66(7):3419-27. doi: 10.1158/0008-5472.CAN-05-1667. Cancer Res. 2006. PMID: 16585163 - Regulation of the G2/M transition by p53.
Taylor WR, Stark GR. Taylor WR, et al. Oncogene. 2001 Apr 5;20(15):1803-15. doi: 10.1038/sj.onc.1204252. Oncogene. 2001. PMID: 11313928 Review. - The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms.
Makeyev AV, Liebhaber SA. Makeyev AV, et al. RNA. 2002 Mar;8(3):265-78. doi: 10.1017/s1355838202024627. RNA. 2002. PMID: 12003487 Free PMC article. Review.
Cited by
- RACK1 identified as the PCBP1-interacting protein with a novel functional role on the regulation of human MOR gene expression.
Nahar-Gohad P, Sultan H, Esteban Y, Stabile A, Ko JL. Nahar-Gohad P, et al. J Neurochem. 2013 Feb;124(4):466-77. doi: 10.1111/jnc.12100. Epub 2012 Dec 26. J Neurochem. 2013. PMID: 23173782 Free PMC article. - Systematic, RNA-interference-mediated identification of mus-101 modifier genes in Caenorhabditis elegans.
Holway AH, Hung C, Michael WM. Holway AH, et al. Genetics. 2005 Mar;169(3):1451-60. doi: 10.1534/genetics.104.036137. Epub 2005 Jan 16. Genetics. 2005. PMID: 15654100 Free PMC article. - Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins.
Yang C, Maiguel DA, Carrier F. Yang C, et al. Nucleic Acids Res. 2002 May 15;30(10):2251-60. doi: 10.1093/nar/30.10.2251. Nucleic Acids Res. 2002. PMID: 12000845 Free PMC article. - Mice deficient in poly(C)-binding protein 4 are susceptible to spontaneous tumors through increased expression of ZFP871 that targets p53 for degradation.
Yan W, Scoumanne A, Jung YS, Xu E, Zhang J, Zhang Y, Ren C, Sun P, Chen X. Yan W, et al. Genes Dev. 2016 Mar 1;30(5):522-34. doi: 10.1101/gad.271890.115. Epub 2016 Feb 25. Genes Dev. 2016. PMID: 26915821 Free PMC article. - Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana.
Lorković ZJ, Barta A. Lorković ZJ, et al. Nucleic Acids Res. 2002 Feb 1;30(3):623-35. doi: 10.1093/nar/30.3.623. Nucleic Acids Res. 2002. PMID: 11809873 Free PMC article.
References
- Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. The p53 network. J Biol Chem. 1998;273:1–4. - PubMed
- Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. - PubMed
- Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J Biol Chem. 1997;272:3129–3132. - PubMed
- Bennett M, Macdonald K, Chan S W, Luzio J P, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous