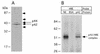CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter - PubMed (original) (raw)
CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter
A M Mikheev et al. Nucleic Acids Res. 2000.
Abstract
In the present study we identified a positive transcriptional element within the rat Ha-ras promoter previously known as Ha-ras response element (HRE) and identified a trans-acting factor that binds HRE sequences in rat mammary cells. To identify the binding protein we employed sequence specific DNA affinity chromatography. Amino acid sequence analysis of the affinity-purified proteins was performed by tandem mass spectroscopy. The results unexpectedly demonstrated that in rat mammary cells CArG box-binding factor A (CBF-A) is the major protein species that bind specifically to the rat and human HRE sequences with high affinity. The affinity of CBF-A binding to HRE was significantly higher than to the CArG box described as a recognition sequence for CBF-A protein. Transient transfection assays using reporter plasmids verified that mutations within the HRE that disrupt binding of CBF-A also reduced the activity of the rat Ha-ras promoter. Despite the fact that the HRE within the Ha-ras promoter resembles a binding site for Ets transcription factors, we did not detect the binding of Ets-related proteins to the rat HRE in BICR-M1Rk cells. We further demonstrated a correlation between the presence of HRE binding activity and induction of Ha-ras mRNA expression following serum stimulation in the mammary carcinoma cell line. Taken together, our results suggest that CBF-A may play an important role in transcriptional regulation of Ha-ras promoter activity during normal mammary cell growth and carcinogenesis.
Figures
Figure 1
EMSA with rat HRE probe and competition experiment with mutant HRE, human HRE (hHRE) and EBS. Competitor DNAs were added in 40-fold molar excess. NE, nuclear extract from BICR-M1Rk. (A) Sequence of oligonucleotides used in the competition experiment. The conserved sequence CCGGAA found in the rat HRE and hHRE and the EBS is boxed. The summary of competition experiment results is on the right. Competition and absence of competition with the HRE probe are indicated by + and –, respectively. (B) EMSA and competition experiment with hHRE and the different mutant oligonucleotides (from 1 to 4) listed on (A). Specific DNA–protein complexes are shown with an arrow. (C) EMSA competition experiment with EBS or rat HRE in the presence of excess rat HRE or EBS respectively. C1 and C2 DNA–protein complexes are indicated. The rat HRE probe without nuclear extract added is not shown.
Figure 2
Wild-type Ha-ras promoter is 3–3.9-fold more active compared to the mutant promoter. BICR-M1Rk cells were transfected with 1 µg of plasmid DNA in serum free conditions and luciferase activity was measured after 8 and 24 h following serum stimulation. Normalization for transfection efficiency was performed as described in Materials and Methods. At the top of the figure is a schematic presentation of the constructs showing wild-type and mutant sequences in the Ha-ras promoter linked to the luciferase gene. Error bars represent standard deviations.
Figure 3
(A) Coomassie Brilliant Blue stained gel following purification of the DNA binding protein by affinity chromatography. Migration of the molecular weight standards is shown on the left. The most abundant species are labeled p42 and p44. Bands of lower intensity which were also subjected to protein identification by micro-HPLC–mass spectrometry are indicated by short arrows. (B) Comparison of radiolabeled, UV crosslinked HRE–protein complexes from whole nuclear extract (NE) with the HRE–p42 complex. The p42 protein was recovered from gel shown on (A). BSA, bovine serum albumin.
Figure 4
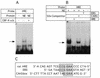
(A) Anti CBF-A antibody completely abrogated protein binding to the rat HRE and human HRE (not shown) probes. EMSA was performed as described in Materials and Methods. Anti CBF-A antibody or normal serum were added as indicated, + and –, respectively. NE, nuclear extract from BICR-M1Rk cells. (B) Comparison of the binding affinity of CBF-A to the rat HRE and CArG box. Competitor oligonucleotides, rat HRE, CArG and EBS were added to the binding reaction in 50-fold molar excess. Specific HRE–protein complex are indicated with arrow. (C) Comparison of rat HRE probe, EBS and CArG box oligonucleotide sequences. Conserved sequence is boxed.
Figure 5
Two protein species (p42 and p44) interact with rat HRE. The western blot was performed with the CBF-A antibody against HRE binding proteins eluted from two sequential affinity columns with 0.1 M KCl step gradient. Aliquots (15 µl) from each dialyzed fraction were used. NE, nuclear extract before loading on the affinity column. The two protein bands, p42 and p44, are indicated by arrows.
Figure 6
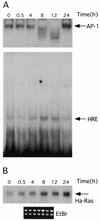
HRE–CBF-A binding activity correlates with Ha-ras mRNA expression. Serum deprived cells were stimulated with 5% calf serum. RNA and nuclear proteins were extracted at indicated time points from the same culture (see Materials and Methods for details). (A) Nuclear proteins were used in EMSA with AP-1 (top, only DNA–protein complexes are shown with arrow) and rat HRE probes (bottom). (B) RNAs extracted from cells at the same time points following serum stimulation were separated on the 1.1% agarose gel, blotted and probed with rat Ha-ras cDNA. Bottom, ethidium bromide staining of the membrane following RNA transfer demonstrates equal RNA loading. For every time point shown on the figure we also performed controls using nuclear extract (A) and RNA (B) from serum deprived cells harvested at a given time. The level of binding activity to the AP-1 and HRE probes and the level of Ha-ras expression did not differ from the zero time points in these samples. For simplicity these controls were removed from the final figure using image analysis software.
Figure 7
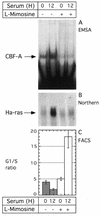
l
-mimosine inhibits CBF-A binding to the rat HRE and expression of the Ha-ras mRNA. Serum deprived cells were stimulated with serum or serum plus
l
-mimosine (200 µg/ml). 12 h later, nuclear proteins and RNA were extracted from the same culture. At each time point, a duplicate cell culture was taken for cell cycle analysis by fluorescence activated cell sorting. (A) EMSA of the CBF-A binding to rat HRE probe. (B) Northern blot analysis of the RNA probed with Ha-ras cDNA. Equal loading was verified by ethidium bromide staining of RNA following transfer to the membrane (not shown). (C) Cell cycle analysis shown the average G1/S ratio from three independent measurements. Error bars represent standard deviations. Open and closed bars, cells untreated and treated with
l
-mimosine, respectively.
Similar articles
- Identification of a novel AU-rich-element-binding protein which is related to AUF1.
Dean JL, Sully G, Wait R, Rawlinson L, Clark AR, Saklatvala J. Dean JL, et al. Biochem J. 2002 Sep 15;366(Pt 3):709-19. doi: 10.1042/BJ20020402. Biochem J. 2002. PMID: 12086581 Free PMC article. - Frequent activation of CArG binding factor-A expression and binding in N-methyl-N-nitrosourea-induced rat mammary carcinomas.
Mikheev AM, Inoue A, Jing L, Mikheeva SA, Li V, Leanderson T, Zarbl H. Mikheev AM, et al. Breast Cancer Res Treat. 2004 Nov;88(1):95-102. doi: 10.1007/s10549-004-1280-5. Breast Cancer Res Treat. 2004. PMID: 15538050 - PEA3 sites within the progression elevated gene-3 (PEG-3) promoter and mitogen-activated protein kinase contribute to differential PEG-3 expression in Ha-ras and v-raf oncogene transformed rat embryo cells.
Su Z, Shi Y, Friedman R, Qiao L, McKinstry R, Hinman D, Dent P, Fisher PB. Su Z, et al. Nucleic Acids Res. 2001 Apr 15;29(8):1661-71. doi: 10.1093/nar/29.8.1661. Nucleic Acids Res. 2001. PMID: 11292838 Free PMC article. - Mechanisms of transcriptional regulation in lymphocyte progenitors: insight from an analysis of the terminal transferase promoter.
Ernst P, Hahm K, Cobb BS, Brown KE, Trinh LA, McCarty AS, Merkenschlager M, Klug CA, Fisher AG, Smale ST. Ernst P, et al. Cold Spring Harb Symp Quant Biol. 1999;64:87-97. doi: 10.1101/sqb.1999.64.87. Cold Spring Harb Symp Quant Biol. 1999. PMID: 11232341 Review. No abstract available. - Identification of base-unpairing region-binding proteins and characterization of their in vivo binding sequences.
Kohwi-Shigematsu T, deBelle I, Dickinson LA, Galande S, Kohwi Y. Kohwi-Shigematsu T, et al. Methods Cell Biol. 1998;53:323-54. doi: 10.1016/s0091-679x(08)60885-7. Methods Cell Biol. 1998. PMID: 9348515 Review. No abstract available.
Cited by
- Identification of S1 proteins B2, C1 and D1 as AUF1 isoforms and their major role as heterogeneous nuclear ribonucleoprotein proteins.
Inoue A, Arao Y, Omori A, Ichinose S, Nishio K, Yamamoto N, Kinoshita Y, Mita S. Inoue A, et al. Biochem J. 2003 Jun 15;372(Pt 3):775-85. doi: 10.1042/BJ20021719. Biochem J. 2003. PMID: 12625834 Free PMC article. - Hnrpab regulates neural development and neuron cell survival after glutamate stimulation.
Sinnamon JR, Waddell CB, Nik S, Chen EI, Czaplinski K. Sinnamon JR, et al. RNA. 2012 Apr;18(4):704-19. doi: 10.1261/rna.030742.111. Epub 2012 Feb 13. RNA. 2012. PMID: 22332140 Free PMC article. - Identification of a novel AU-rich-element-binding protein which is related to AUF1.
Dean JL, Sully G, Wait R, Rawlinson L, Clark AR, Saklatvala J. Dean JL, et al. Biochem J. 2002 Sep 15;366(Pt 3):709-19. doi: 10.1042/BJ20020402. Biochem J. 2002. PMID: 12086581 Free PMC article. - Characterization of nuclear factors modulating the apolipoprotein D promoter during growth arrest: implication of PARP-1, APEX-1 and ERK1/2 catalytic activities.
Levros LC Jr, Do Carmo S, Edouard E, Legault P, Charfi C, Rassart E. Levros LC Jr, et al. Biochim Biophys Acta. 2010 Sep;1803(9):1062-71. doi: 10.1016/j.bbamcr.2010.04.011. Epub 2010 May 21. Biochim Biophys Acta. 2010. PMID: 20493910 Free PMC article. - Hnrnpab regulates neural cell motility through transcription of Eps8.
Lampasona AA, Czaplinski K. Lampasona AA, et al. RNA. 2019 Jan;25(1):45-59. doi: 10.1261/rna.067413.118. Epub 2018 Oct 12. RNA. 2019. PMID: 30314980 Free PMC article.
References
- Downward J. (1997) Curr. Biol., 7, R258–R260. - PubMed
- Lloyd A.C. (1998) Curr. Opin. Genet. Dev., 8, 43–48. - PubMed
- Finney R.E. and Bishop,J.M. (1993) Science, 260, 1524–1527. - PubMed
- Yaginuma Y., Yamashita,K., Kuzumaki,N., Fujita,M. and Shimizu,T. (1992) Gynecol. Oncol., 46, 45–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials