5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence - PubMed (original) (raw)
5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence
B Zhu et al. Nucleic Acids Res. 2000.
Abstract
A 1468 bp cDNA coding for the chicken homolog of the human MBD4 G/T mismatch DNA glycosylase was isolated and sequenced. The derived amino acid sequence (416 amino acids) shows 46% identity with the human MBD4 and the conserved catalytic region at the C-terminal end (170 amino acids) has 90% identity. The non-conserved region of the avian protein has no consensus sequence for the methylated DNA binding domain. The recombinant proteins from human and chicken have G/T mismatch as well as 5-methylcytosine (5-MeC) DNA glycosylase activities. When tested by gel shift assays, human recombinant protein with or without the methylated DNA binding domain binds equally well to symmetrically, hemimethylated DNA and non-methylated DNA. However, the enzyme has only 5-MeC DNA glycosylase activity with the hemimethylated DNA. Footprinting of human MBD4 and of an N-terminal deletion mutant with partially depurinated and depyrimidinated substrate reveal a selective binding of the proteins to the modified substrate around the CpG. As for 5-MeC DNA glycosylase purified from chicken embryos, MBD4 does not use oligonucleotides containing mCpA, mCpT or mCpC as substrates. An mCpG within an A+T-rich oligonucleotide is a much better substrate than an A+T-poor sequence. The K:(m) of human MBD4 for hemimethylated DNA is approximately 10(-7) M with a V:(max) of approximately 10(-11) mol/h/microgram protein. Deletion mutations show that G/T mismatch and 5-MeC DNA glycosylase are located in the C-terminal conserved region. In sharp contrast to the 5-MeC DNA glycosylase isolated from the chicken embryo DNA demethylation complex, the two enzymatic activities of MBD4 are strongly inhibited by RNA. In situ hybridization with antisense RNA indicate that MBD4 is only located in dividing cells of differentiating embryonic tissues.
Figures
Figure 1
Comparison of the deduced amino acid sequence of the chicken MBD4 (accession no. AF257107) with the human MBD4 (accession no. AF072250). The boxed sequence corresponds to the conserved region of mammalian mCpG binding proteins (methylated DNA binding domain).
Figure 2
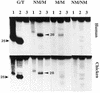
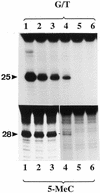
Substrate specificity of 5-MeC DNA glycosylase. The upper panel is the reactions with human MBD4 and the lower panel, the reactions with the chicken homolog of human MBD4. Enzymatic reactions were carried out as described in Materials and Methods. The arrows point to the correct positions of the cleavage products which are 25 and 28 nt, respectively, for the G/T mismatch glycosylase and 5-MCDG (oligos A and B, respectively, Table 1). G/T, substrate with a G/T mismatch; NM, non-methylated DNA strand; M, methylated DNA strand. In lanes 1, the reaction product was denatured before loading onto the gel whereas in lanes 2, reaction products were treated with 0.1 M NaOH at 95°C for 10 min before loading onto the sequencing gel. In lanes 3, the substrate was incubated with BSA only and was denatured prior to loading onto the DNA sequencing gel (20% polyacrylamide–urea).
Figure 3
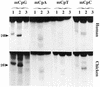
Sequence specificity of 5-MeC DNA glycosylase. The upper panel represents the activity for human MBD4 and the lower panel the activity for the chicken homolog of MBD4. In lanes 1 the reaction product is denatured at 95°C before loading (this shows possible specific nicks at the 3′ position of the abasic sugar). In lanes 2 the reaction product is treated for 10 min at 95°C in 0.1 M NaOH before loading (this reaction removes the abasic sugar from the oligonucleotide, giving a shift in the migration of the band). In lanes 3 a blank is denatured at 95°C for 5 min before loading. The arrow points to the position of the reaction product which is 28 nt long for mCpG (see also Table 1). All substrates were hemimethylated and contained either one mCpG, mCpA, mCpT or mCpC (oligos B, C, D and E, respectively, Table 1).
Figure 4
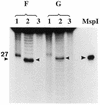
Influence of the A+T content of the oligonucleotide substrates on the activity of 5-MeC DNA glycosylase. Each assay had 50 ng of human recombinant MBD4 per 50 µl incubation mixture. Incubation was for 30 min at 37°C. Oligo F has 92% A+T whereas oligo G has 46% A+T. In lanes 1 the product of the reactions was denatured before loading onto the 20% acrylamide sequencing gel, whereas in lanes 2 the product of reaction was treated for 5 min at 95°C with 0.1 M NaOH before loading (cleavage and removal of the abasic sugar). Lanes 3 are the controls incubated without glycosylase. MspI is the 27 bp size marker resulting from cleavage of the substrate with _Msp_I.
Figure 5
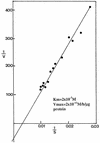
Determination of the _K_m and _V_max for hemimethylated DNA substrate incubated with recombinant human MBD4. Incubation conditions were as outlined in Materials and Methods. Reaction product was analyzed on a 20% urea–polyacrylamide DNA sequencing gel. Upon autoradiography, bands were cut out and counted for radioactivity. Controls were carried out with the DNA substrate only. An area corresponding to the reaction product was cut out, counted and subtracted from the test values. Results are expressed as a Lineweaver–Burk plot.
Figure 6
Deletion mutations of human MBD4. (A) The N- and C-terminal deletion mutants. FL represents the full-length protein. The relative activity of G/T mismatch and 5-MeC DNA glycosylase are indicated by +++, ++, +, ± and –. The radioactive bands corresponding to the specific reaction products were cut out from the gel and counted for radioactivity. The ratio of the two enzyme activities is given. (B) Silver stained gel of the mutant enzymes isolated on a Ni-NTA–agarose column.
Figure 7
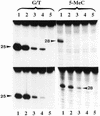
Analysis of N- and C-terminal mutants for G/T mismatch and 5-MeC DNA glycosylase activities. The enzyme preparations shown in Figure 6B were assayed for G/T mismatch and 5-MeC DNA glycosylase activities. The reaction product was treated with NaOH and separated on a 20% urea–polyacrylamide DNA sequencing gel. Lanes 1–5 correspond to full-length protein, ΔN273, ΔN378, ΔN433 and ΔC48, respectively, and lane 6 is a blank. The arrowheads point to the positions of the reaction products which are 25 and 28 nt long for the G/T mismatch glycosylase and 5-MCDG, respectively.
Figure 8
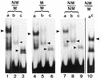
Gel shift assay of recombinant MBD4 and its mutants with hemimethylated substrate (NM/M), symmetrically-methylated substrate (M/M) and non-methylated substrate NM/NM. Lanes a, human MBD4 (MW 60 000 Da); lanes b, mutant ΔN273; lanes c, mutant ΔN378. Lane ac is the chicken wild-type MBD4 (MW 40 000 Da). Gel shifts were carried out as outlined in Materials and Methods and the reaction product was analyzed in a 5% native polyacrylamide gel. The arrowheads point to the specific complex of the glycosylase and its mutants with the DNA substrate (see also Fig. 9).
Figure 9
Which of the two gel shift bands a and b seen with the wild-type MBD4 contains 5-MCDG activity? Lane 1 is a gel shift assay of the human MBD4 with the hemimethylated substrate. Bands a and b are the protein DNA complex and c is the free DNA. Gel shift assays were carried out as outlined in Materials and Methods. Lanes 2–7 are the products of the reactions from bands a, b and c analyzed on a 20% acrylamide DNA sequencing gel. In the lanes marked –, the oligonucleotides extracted from bands a, b and c were denatured and analyzed on the sequencing gel whereas for the lanes marked with a +, the product of the reaction was subjected to alkaline hydrolysis before analyzing on the DNA sequencing gel. The size of the correct reaction product of 5-MCDG (arrowhead) is 28 nt.
Figure 10
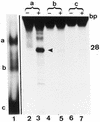
Missing contact probing of MBD4 and its mutant Δ273 with partially depurinated and depyrimidinated hemimethylated substrate. Experiment was carried out as outlined in Materials and Methods. C is the control DNA substrate partially depurinated (A+G) or depyrimidinated (C+T), B is the modified substrate bound to the enzyme in the gel shift assay and F is the unbound free modified substrate migrating at the bottom of the gel (Fig. 9). The substrate was hemimethylated. The mCpG is located on the lower strand and the upper strand is non-methylated. Numbers in the lanes refer to the nucleotide positions shown in the DNA sequence of Figure 11 (oligo B).
Figure 11
Trend analysis of the results shown in Figure 10. From the pool of randomly modified oligos (partial depurination and depyrimidination) MBD4 and its mutant have a selective affinity for some abasic sites. The stronger the affinity to such sites, the stronger are the signals in the B lanes when compared with the C and F lanes. The strongest signals are marked with a closed circle and the weaker one with an open circle, M represents 5-MeC. The numbers indicate the nucleotide position in the DNA substrate (oligo B) which is 50 bp long.
Figure 12
Effect of increasing concentrations of synthetic oligoribonucleotides on the activity of G/T mismatch and 5-MeC DNA glycosylases. In the upper panels the enzyme (human MBD4) was incubated with 0, 0.25, 0.5 and 1 µg of the oligo 5′-GUGACCGGAGC-3′ complementary to the target sequence whereas the lower panels are the enzyme (chicken MBD4) incubated with 0, 0.25, 0.5 and 1 µg of 5′-GCUCCGGUCAC-3′ complementary to the opposite strand. Lane 5 is the blank incubated without protein. The reaction product was subjected to alkaline hydrolysis and analyzed on a 20% acrylamide sequencing gel. The arrowheads point to the correct position of the cleavage products.
Figure 13
The chicken MBD4 RNA is only transcribed in mitotically active tissues. (A) Section through the eye of a 5-day-old chicken embryo. Transcripts are present in the neuroepithelia of the optical cup (oc) but not in the cells of the lens (arrows) producing the crystalline. (C) Section adjacent to (A) probed with sense RNA (negative control). (B and D) DAPI staining of the corresponding sections.
Similar articles
- 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex.
Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost JP. Zhu B, et al. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5135-9. doi: 10.1073/pnas.100107597. Proc Natl Acad Sci U S A. 2000. PMID: 10779566 Free PMC article. - A chicken embryo protein related to the mammalian DEAD box protein p68 is tightly associated with the highly purified protein-RNA complex of 5-MeC-DNA glycosylase.
Jost JP, Schwarz S, Hess D, Angliker H, Fuller-Pace FV, Stahl H, Thiry S, Siegmann M. Jost JP, et al. Nucleic Acids Res. 1999 Aug 15;27(16):3245-52. doi: 10.1093/nar/27.16.3245. Nucleic Acids Res. 1999. PMID: 10454630 Free PMC article. - Mechanisms of DNA demethylation in chicken embryos. Purification and properties of a 5-methylcytosine-DNA glycosylase.
Jost JP, Siegmann M, Sun L, Leung R. Jost JP, et al. J Biol Chem. 1995 Apr 28;270(17):9734-9. doi: 10.1074/jbc.270.17.9734. J Biol Chem. 1995. PMID: 7730351 - [Structure basis of versatile base recognition of MBD4].
Ariyoshi M, Otani J, Shirakawa M. Ariyoshi M, et al. Yakugaku Zasshi. 2015;135(1):3-9. doi: 10.1248/yakushi.14-00202-1. Yakugaku Zasshi. 2015. PMID: 25743892 Review. Japanese. - Structural and mutation studies of two DNA demethylation related glycosylases: MBD4 and TDG.
Hashimoto H. Hashimoto H. Biophysics (Nagoya-shi). 2014 Oct 18;10:63-8. doi: 10.2142/biophysics.10.63. eCollection 2014. Biophysics (Nagoya-shi). 2014. PMID: 27493500 Free PMC article. Review.
Cited by
- No evidence for AID/MBD4-coupled DNA demethylation in zebrafish embryos.
Shimoda N, Hirose K, Kaneto R, Izawa T, Yokoi H, Hashimoto N, Kikuchi Y. Shimoda N, et al. PLoS One. 2014 Dec 23;9(12):e114816. doi: 10.1371/journal.pone.0114816. eCollection 2014. PLoS One. 2014. PMID: 25536520 Free PMC article. - Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites.
Bellacosa A, Drohat AC. Bellacosa A, et al. DNA Repair (Amst). 2015 Aug;32:33-42. doi: 10.1016/j.dnarep.2015.04.011. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 26021671 Free PMC article. Review. - Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice.
Kemmerich K, Dingler FA, Rada C, Neuberger MS. Kemmerich K, et al. Nucleic Acids Res. 2012 Jul;40(13):6016-25. doi: 10.1093/nar/gks259. Epub 2012 Mar 24. Nucleic Acids Res. 2012. PMID: 22447450 Free PMC article. - Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes.
D'Alessio AC, Weaver IC, Szyf M. D'Alessio AC, et al. Mol Cell Biol. 2007 Nov;27(21):7462-74. doi: 10.1128/MCB.01120-07. Epub 2007 Aug 20. Mol Cell Biol. 2007. PMID: 17709385 Free PMC article. - TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model.
Ge L, Zhang RP, Wan F, Guo DY, Wang P, Xiang LX, Shao JZ. Ge L, et al. Mol Cell Biol. 2014 Mar;34(6):989-1002. doi: 10.1128/MCB.01061-13. Epub 2014 Jan 6. Mol Cell Biol. 2014. PMID: 24396069 Free PMC article.
References
- Russo V.E.A., Martienssen,R.S. and Riggs,A.D. (1986) Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Laird P.W. (2000) In Jones,P.A. and Vogt,P.K. (eds), DNA Methylation and Cancer. Springer Verlag, pp. 119–134.
- Jost J.P., Oakeley,E.J. and Schwarz,S. (1999) In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosyl Methionine Dependent Methyltransferases: Structures and Functions. World Scientific.
- Jost J.P., Siegmann,M., Thiry,S., Jost,Y.C., Benjamin,D. and Schwarz,S. (1999) FEBS Lett., 449, 251–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous