Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells - PubMed (original) (raw)
Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells
W H Tong et al. EMBO J. 2000.
Abstract
Iron-sulfur (Fe-S) clusters are cofactors found in many proteins that have important redox, catalytic or regulatory functions. In mammalian cells, almost all known Fe-S proteins are found in the mitochondria, but at least one is found in the cytosol. Here we report cloning of the human homologs to IscU and NifU, iron-binding proteins that play a critical role in Fe-S cluster assembly in bacteria. In human cells, alternative splicing of a common pre-mRNA results in synthesis of two proteins that differ at the N-terminus and localize either to the cytosol (IscU1) or to the mitochondria (IscU2). Biochemical analyses demonstrate that IscU proteins specifically associate with IscS, a cysteine desulfurase that is proposed to sequester inorganic sulfur for Fe-S cluster assembly. Protein complexes containing IscU and IscS can be found in the mitochondria as well as in the cytosol, implying that Fe-S cluster assembly takes place in multiple subcellular compartments in mammalian cells. The possible roles of the IscU proteins in mammalian cells and the potential implications of compartmentalization of Fe-S cluster assembly are discussed.
Figures
Fig. 1. Molecular cloning and sequence analysis of human iscU. (A) Predicted amino acid sequences of two human IscU isoforms compared with yeast Isu1 and Isu2. The cDNA sequences of human IscU were obtained by EST database analysis and 5′ RACE experiments. Sequence analysis of 5′ RACE products, EST and genome databases has revealed polymorphisms at codon 7 (G or F) and codon 12 (V or A) in the N-terminus of IscU2. Only one variant is shown here. (B) Genomic sequence of the 5′ end of human iscU obtained by PCR and EST database analysis.
Fig. 2. Alternative splicing of iscU pre-mRNA results in two isoforms with different predicted N-terminal sequences. (A) Schematic representation of iscU1 and iscU2 5′ cDNA ends. Nested PCR of a 5′ RACE fragment was carried out using a cap-binding primer, in combination with iscU gene-specific primers. Sequence analysis indicated that iscU1 and iscU2 are alternative splice products. (B) Nested PCR of a 5′ RACE fragment was carried out using a 3′ primer within exon III in combination with 5′ primers specific to either exon IA or exon IB. Southern blot analysis was carried out using exon IA, exon IB or intron IB probe. Arrowheads denote bands of the appropriate sizes as expected from iscU1 and iscU2 mRNA sequences. Asterisks denote bands containing intron sequence and are derived from splicing intermediates. (C) Northern blot analysis of poly(A)+ RNA from RD4 cells and human heart (Clontech) was carried out using exon IA, exon IB or intron IB probe. Arrowheads denote bands of the appropriate sizes as expected from iscU1 and iscU2 mRNA sequences. Asterisks denote splicing intermediates. (D) Human multiple tissue northern blotting (Clontech) revealed that iscU, iscS and iscA are all expressed predominantly in heart and skeletal muscle.
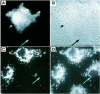
Fig. 3. IscU1 is localized to cytosol and nucleus, whereas IscU2 is localized to mitochondria. COS cells transfected with HA-tagged IscU1 were stained with monoclonal anti-HA antibodies and analyzed by immunofluorescence microscopy (A) and phase contrast microscopy (B). (C) COS cells transfected with myc-tagged IscU2 were stained with monoclonal anti-myc antibodies and analyzed by immunofluorescence microscopy. (D) Mitochondrial staining by the mitochondrial marker rhodamine 123 of transfected COS cells shown in (C). Short arrows denote transfected cells, whereas long arrows denote untransfected cells.
Fig. 4. IscU2 protein is rapidly processed and becomes associated with an ∼47 kDa protein upon maturation. (A) Schematic representation of the full-length IscU2 construct and IscU2Δ35, a construct that lacks sequence 5′ of the putative processing site. (B) COS cells transfected with a construct encoding a full-length myc-tagged IscU2 were pulse labeled with [35S]methionine for 3 min, and chased for 0, 15, 60 and 300 min, respectively. Immunoprecipitation with a monoclonal anti-myc antibody indicated a two-step processing of the IscU2 protein.In vitro translation of myc-tagged IscU2Δ35 generated a protein that co-migrated with the processed IscU2 in transfected COS cells. At the 60 and 300 min time points, a 47 kDa band (arrowhead) was co-immunoprecipitated with IscU.
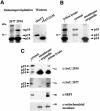
Fig. 5. Endogenous IscU proteins are present in the cytosol and in the mitochondria of RD4 cells. (A) Immunoprecipitation experiments revealed multiple forms of endogenous IscU proteins in RD4 cells. Immunoprecipitations were carried out using affinity-purified anti-IscU peptide antibodies 2934 and 2677. Western blot analyses of recombinant IscU1 (rIscU1) and recombinant IscU2Δ35 (rIscU2Δ35) are shown for comparison. (B) Immunoprecipitation of endogenous IscU proteins in different subcellular fractions of RD4 cell lysates using anti-IscU 2934. (C) Western blot analyses of endogenous IscU proteins in different subcellular fractions of RD4 cell lysates. Western blot analyses of mitochondrial aconitase and cytosolic IRP1 are shown for comparison.
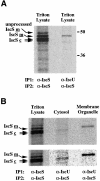
Fig. 6. Co-immunoprecipitation of IscU and IscS proteins in the cytosol and in the mitochondria of RD4 cells. (A) Coimmuno-precipitation of mitochondrial IscS with IscU in RD4 Triton lysates. The mitochondrial IscU–IscS complex was first immunoprecipitated from [35S]methionine-labeled RD4 cell lysate using either α-IscS or α-IscU 2934. Following dissociation of the immunoprecipitates from the antibodies with SDS–DTT and heating, a second immuno precipitation (recapture) using α-IscS was carried out. (B) Cytosolic IscU1 co-immunoprecipitates with one of the cytosolic IscS proteins (IscS
c
), and mitochondrial IscU2 co-immunoprecipitates with mitochondrial IscS (IscS
m
) in RD4 cell lysates (upper panel). The lower panel shows the lanes containing the cytosolic and membrane organelle fractions at high contrast to allow better visualization and alignment of the cytosolic IscS band.
Similar articles
- The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes.
Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT, Hunt JF. Goldsmith-Fischman S, et al. J Mol Biol. 2004 Nov 19;344(2):549-65. doi: 10.1016/j.jmb.2004.08.074. J Mol Biol. 2004. PMID: 15522304 - Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron-sulfur cluster assembly.
Kato S, Mihara H, Kurihara T, Takahashi Y, Tokumoto U, Yoshimura T, Esaki N. Kato S, et al. Proc Natl Acad Sci U S A. 2002 Apr 30;99(9):5948-52. doi: 10.1073/pnas.082123599. Epub 2002 Apr 23. Proc Natl Acad Sci U S A. 2002. PMID: 11972033 Free PMC article. - A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans.
You D, Wang L, Yao F, Zhou X, Deng Z. You D, et al. Biochemistry. 2007 May 22;46(20):6126-33. doi: 10.1021/bi602615k. Epub 2007 May 1. Biochemistry. 2007. PMID: 17469805 - Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer.
Fontecave M, Ollagnier-de-Choudens S. Fontecave M, et al. Arch Biochem Biophys. 2008 Jun 15;474(2):226-37. doi: 10.1016/j.abb.2007.12.014. Epub 2007 Dec 28. Arch Biochem Biophys. 2008. PMID: 18191630 Review. - Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases.
Lill R, Mühlenhoff U. Lill R, et al. Annu Rev Biochem. 2008;77:669-700. doi: 10.1146/annurev.biochem.76.052705.162653. Annu Rev Biochem. 2008. PMID: 18366324 Review.
Cited by
- A Northwestern blotting approach for studying iron regulatory element-binding proteins.
Popovic Z, Templeton DM. Popovic Z, et al. Mol Cell Biochem. 2005 Jan;268(1-2):67-74. doi: 10.1007/s11010-005-3167-0. Mol Cell Biochem. 2005. PMID: 15724439 - Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly.
Frazzon AP, Ramirez MV, Warek U, Balk J, Frazzon J, Dean DR, Winkel BS. Frazzon AP, et al. Plant Mol Biol. 2007 Jun;64(3):225-40. doi: 10.1007/s11103-007-9147-x. Epub 2007 Apr 7. Plant Mol Biol. 2007. PMID: 17417719 - A novel eukaryotic factor for cytosolic Fe-S cluster assembly.
Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE. Roy A, et al. EMBO J. 2003 Sep 15;22(18):4826-35. doi: 10.1093/emboj/cdg455. EMBO J. 2003. PMID: 12970194 Free PMC article. - Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein.
Ding H, Clark RJ. Ding H, et al. Biochem J. 2004 Apr 15;379(Pt 2):433-40. doi: 10.1042/BJ20031702. Biochem J. 2004. PMID: 14720122 Free PMC article.
References
- Agar J.N., Zheng,L., Cash,V.L., Dean,D.R. and Johnson,M.K. (2000) Role of IscU protein in iron–sulfur cluster biosynthesis: IscS-mediated assembly of a [Fe2S2] cluster in IscU. J. Am. Chem. Soc., 122, 2136–2137.
- Askwith C. and Kaplan,J. (1998) Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci., 23, 135–138. - PubMed
- Babcock M., de Silva,D., Oaks,R., Davis-Kaplan,S., Jiralerspong,S., Montermini,L., Pandolfo,M. and Kaplan,J. (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science, 276, 1709–1712. - PubMed
- Beinert H. and Holm,R.H. (1997) Iron–sulfur clusters: nature’s modular, multipurpose structures. Science, 277, 653–659. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous