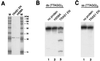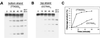Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1 - PubMed (original) (raw)
Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1
T Anzai et al. Mol Cell Biol. 2001 Jan.
Abstract
The telomere of the silkworm Bombyx mori consists of (TTAGG/CCTAA)(n) repeats and harbors a large number of telomeric repeat-specific non-long terminal repeat retrotransposons, such as TRAS1 and SART1. To understand how these retrotransposons recognize and integrate into the telomeric repeat in a sequence-specific manner, we expressed the apurinic-apryrimidinic endonuclease-like endonuclease domain of TRAS1 (TRAS1 EN), which is supposed to digest the target DNA, and characterized its enzymatic properties. Purified TRAS1 EN could generate specific nicks on both strands of the telomeric repeat sequence between T and A of the (TTAGG)(n) strand (bottom strand) and between C and T of the (CCTAA)(n) strand (top strand). These sites are consistent with insertion sites expected from the genomic structure of boundary regions of TRAS1. Time course studies of nicking activities on both strands revealed that the cleavages on the bottom strand preceded those on the top strand, supporting the target-primed reverse transcription model. TRAS1 EN could cleave the telomeric repeats specifically even if it was flanked by longer tracts of nontelomeric sequence, indicating that the target site specificity of the TRAS1 element was mainly determined by its EN domain. Based on mutation analyses, TRAS1 EN recognizes less than 10 bp around the initial cleavage site (upstream 7 bp and downstream 3 bp), and the GTTAG sequence especially is essential for the cleavage reaction on the bottom strand (5'. TTAGGTT downward arrow AGG. 3'). TRAS1 EN, the first identified endonuclease digesting telomeric repeats, may be used as a genetic tool to shorten the telomere in insects and some other organisms.
Figures
FIG. 1
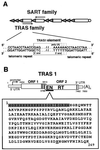
Telomeric sequence-specific retrotransposons of the silkworm, B. mori. (A) Chromosome structure of B. mori. The shaded triangle indicates the telomeric repeat sequence 5′-(TTAGG)n/5′-(CCTAA)n. Open boxes indicate retrotransposons TRAS and SART, which are inserted into the telomeric repeat in opposite directions. Arrows show the directions of transcription by these elements. The 5′ and 3′ junction regions between TRAS1 and the telomeric repeat are shown below. The precise 3′ end of TRAS1 has not been determined. Dotted boxes indicate possible junction sites. (B) Structure of TRAS1. TRAS1 is inserted with its poly(A) tail facing toward the centromere. The ORFs of TRAS1 are depicted by open boxes, and the positions of a promoter, EN domain, and RT domain are indicated. The vertical lines near the C terminus of both ORFs represent the cysteine-histidine motifs. The amino acid sequence of TRAS1 EN expressed in E. coli is shown below. The N-terminal His tag sequence derived from pET16b is shaded. The putative active site (His-258) for endonuclease activity, which was changed to Ala in the mutant experiment, is underlined.
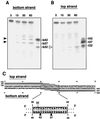
FIG. 2
Expression and nicking activities of TRAS1 EN. (A) Purified TRAS1 EN and mutant protein H258A were separated on an SDS–8% PAGE gel and stained with Coomassie blue. Identical bands could be observed at 30.3 kDa. Lanes M, size markers. (B) Activity for double-stranded oligonucleotides. Telomeric G-strand (bottom strand) was end labeled and annealed to the complementary nonlabeled C-strands (top strand). Double-stranded (TTAGG)5 was treated with no protein (lane 1), 0.2 μg of H258A mutant protein (lane 2), and 0.2 μg of TRAS1 EN (lane 3) at 25°C for 60 min. The resulting fragments were separated on a 28% denaturing gel. (C) Activity for single-stranded oligonucleotides. Single-stranded (TTAGG)5 was prepared and incubated with no protein (lane 1) and with TRAS1 EN protein (lane 2). ds, double-stranded DNA; ss, single-stranded DNA.
FIG. 3
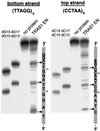
Site-specific cleavage on both strands of telomeric repeats by TRAS1 EN. The 32P-end-labeled (TTAGG)5 (bottom strand) and (CCTAA)6 (top strand) were annealed with the complementary oligonucleotides. Reactions with TRAS1 EN were conducted as in Fig. 2 except that twice the amount of TRAS1 EN (0.4 μg) was added. Such an excess of TRAS1 EN caused nonspecific bands, which are useful for identifying the band positions (see text). Various lengths of 32P-labeled oligonucleotides (dG10, dG12, dG15, dG17, dC10, dC12, dC15, and dC17) were loaded as molecular standards (Materials and Methods). The major nicking sites by TRAS1 EN are shown by arrows on the right.
FIG. 4
Kinetics of the cleavage reaction of TRAS1 EN. (A and B) Time course of nicking activities on the each strand of telomeres. TRAS1 EN (0.2 μg) was incubated with double-stranded substrates. Incubation time (in minutes) at 25°C is indicated on the top. Note that bottom-strand cleavage precedes top-strand cleavage. dG12, dG17, dC12, and dC17 were used as size markers. (C) Quantitation of the cleavage reactions shown in panels A and B. The 17-bp cleaved product (*5′-[TTAGG]3-TT↓-3′) in the bottom-strand reaction was quantified with the BAS system. Similarly, the 12-bp product (*5′-[CCTAA]2-CC↓-3′) in the top-strand reaction was measured. The percentage of nicked substrate was plotted against incubation time.
FIG. 5
Target-specific cleavage of a long DNA substrate by TRAS1 EN. (A and B) A 75-bp-long DNA substrate contained a 15-bp telomere sequence (boxed region in C) flanked by nontelomeric sequences at both ends. The entire sequence of 75 bp is shown in panel C. The end-labeled 75-bp DNA was annealed with its complementary strand and cleaved with TRAS1 EN under the same conditions as in Fig. 2. Incubation time (in minutes) at 25°C is indicated on the top. Various lengths of end-labeled oligonucleotides (b32, b37, b42, t32, t37, and t42) were loaded as exact size markers as shown in panel C. (C) Schematic representation of the cleavage sites on both the top and bottom strands of the 75-bp substrate. Nucleotides are counted from the 5′ end of each strand. The solid arrowheads indicate cleavage sites on the bottom sequence. The open arrowhead marks the cleavage site on the top strand. A putative cleavage pattern on the double-stranded DNA is shown by a dotted line.
FIG. 6
Endonucleolytic activities of TRAS1 EN for mutated telomeric repeats. Each nucleotide of the TTAGG unit of the bottom strand was changed individually to a cytosine residue. Treatment with TRAS1 EN protein was performed as described in the legend to Fig. 2. Mutated nucleotides are underlined. EN: −, without TRAS1 EN; +, with TRAS1 EN.
FIG. 7
Definition of the bases involved in the cleavage reaction of TRAS1 EN. Treatment with TRAS1 EN protein was performed as described for Fig. 2. Each base from −7 to +8, where 0 is the cleavage site of the 12-bp cleaved product, in the 25-bp (TTAGG)5 bottom strand was systematically changed to a cytosine (boxed). The cleavage patterns around the 12-bp cleaved product in the gel are also shown. cont., control. The 12-bp band in each reaction was quantified, and its intensity relative to the control is shown on the right (control = 100). Each value represents the average, and error bars represent the standard error; both values were obtained from six independent experiments.
Similar articles
- Structural and phylogenetic analysis of TRAS, telomeric repeat-specific non-LTR retrotransposon families in Lepidopteran insects.
Kubo Y, Okazaki S, Anzai T, Fujiwara H. Kubo Y, et al. Mol Biol Evol. 2001 May;18(5):848-57. doi: 10.1093/oxfordjournals.molbev.a003866. Mol Biol Evol. 2001. PMID: 11319268 - Structural analysis of TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm, Bombyx mori.
Okazaki S, Ishikawa H, Fujiwara H. Okazaki S, et al. Mol Cell Biol. 1995 Aug;15(8):4545-52. doi: 10.1128/MCB.15.8.4545. Mol Cell Biol. 1995. PMID: 7623845 Free PMC article. - Transcription analysis of the telomeric repeat-specific retrotransposons TRAS1 and SART1 of the silkworm Bombyx mori.
Takahashi H, Fujiwara H. Takahashi H, et al. Nucleic Acids Res. 1999 May 1;27(9):2015-21. doi: 10.1093/nar/27.9.2015. Nucleic Acids Res. 1999. PMID: 10198435 Free PMC article. - Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori.
Fujiwara H, Osanai M, Matsumoto T, Kojima KK. Fujiwara H, et al. Chromosome Res. 2005;13(5):455-67. doi: 10.1007/s10577-005-0990-9. Chromosome Res. 2005. PMID: 16132811 Review. - [Cytogenetics and application in domesticated silkworm Bombyx mori].
Li W, Ge FL, Ye DP, Lei JH, Huang M. Li W, et al. Yi Chuan. 2006 Sep;28(9):1167-72. doi: 10.1360/yc-006-1167. Yi Chuan. 2006. PMID: 16963430 Review. Chinese.
Cited by
- The molecular biology of human herpesvirus-6 latency and telomere integration.
Arbuckle JH, Medveczky PG. Arbuckle JH, et al. Microbes Infect. 2011 Aug;13(8-9):731-41. doi: 10.1016/j.micinf.2011.03.006. Epub 2011 Mar 31. Microbes Infect. 2011. PMID: 21458587 Free PMC article. Review. - At the Beginning of the End and in the Middle of the Beginning: Structure and Maintenance of Telomeric DNA Repeats and Interstitial Telomeric Sequences.
Aksenova AY, Mirkin SM. Aksenova AY, et al. Genes (Basel). 2019 Feb 5;10(2):118. doi: 10.3390/genes10020118. Genes (Basel). 2019. PMID: 30764567 Free PMC article. Review. - The telomere-specific non-LTR retrotransposons SART1 and TRAS1 are suppressed by Piwi subfamily proteins in the silkworm, Bombyx mori.
Tatsuke T, Sakashita K, Masaki Y, Lee JM, Kawaguchi Y, Kusakabe T. Tatsuke T, et al. Cell Mol Biol Lett. 2010;15(1):118-33. doi: 10.2478/s11658-009-0038-9. Epub 2009 Nov 27. Cell Mol Biol Lett. 2010. PMID: 19943120 Free PMC article. - Characterization of the sequence specificity of the R1Bm endonuclease domain by structural and biochemical studies.
Maita N, Aoyagi H, Osanai M, Shirakawa M, Fujiwara H. Maita N, et al. Nucleic Acids Res. 2007;35(12):3918-27. doi: 10.1093/nar/gkm397. Epub 2007 May 30. Nucleic Acids Res. 2007. PMID: 17537809 Free PMC article. - Repeat-induced point mutation and the population structure of transposable elements in Microbotryum violaceum.
Hood ME, Katawczik M, Giraud T. Hood ME, et al. Genetics. 2005 Jul;170(3):1081-9. doi: 10.1534/genetics.105.042564. Epub 2005 May 23. Genetics. 2005. PMID: 15911572 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials