RNA interference is mediated by 21- and 22-nucleotide RNAs - PubMed (original) (raw)
RNA interference is mediated by 21- and 22-nucleotide RNAs
S M Elbashir et al. Genes Dev. 2001.
Abstract
Double-stranded RNA (dsRNA) induces sequence-specific posttranscriptional gene silencing in many organisms by a process known as RNA interference (RNAi). Using a Drosophila in vitro system, we demonstrate that 21- and 22-nt RNA fragments are the sequence-specific mediators of RNAi. The short interfering RNAs (siRNAs) are generated by an RNase III-like processing reaction from long dsRNA. Chemically synthesized siRNA duplexes with overhanging 3' ends mediate efficient target RNA cleavage in the lysate, and the cleavage site is located near the center of the region spanned by the guiding siRNA. Furthermore, we provide evidence that the direction of dsRNA processing determines whether sense or antisense target RNA can be cleaved by the siRNA-protein complex.
Figures
Figure 1
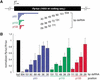
Double-stranded RNA as short as 38 bp can mediate RNAi. (A) Graphic representation of dsRNAs used for targeting _Pp_-luc mRNA. Three series of blunt-ended dsRNAs covering a range of 29–504 bp were prepared. The position of the first nucleotide of the sense strand of the dsRNA is indicated relative to the start codon of _Pp_-luc mRNA (p1). (B) RNA interference assay (Tuschl et al. 1999). Ratios of target _Pp_-luc to control _Rr_-luc activity were normalized to a buffer control (black bar). DsRNAs (5 nM) were preincubated in Drosophila lysate at 25°C for 15 min before the addition of 7-methyl-guanosine-capped _Pp_-luc and _Rr_-luc mRNAs (∼50 pM). The incubation was continued for another hour and then analyzed by the dual luciferase assay (Promega). The data are the average from at least four independent experiments ±S.D.
Figure 2
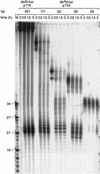
A 29-bp dsRNA is only slowly processed to 21–23-nt fragments. Time course of 21–23-mer formation from processing of internally 32P-labeled dsRNAs (5 nM) in the Drosophila lysate. The length and source of the dsRNA are indicated. An RNA size marker (M) has been loaded in the left lane, and the fragment sizes are indicated. Double bands at time zero are caused by incompletely denatured dsRNA.
Figure 3
Mapping of sense and antisense target RNA cleavage sites. (A) Denaturing gel electrophoresis of the stable 5′ cleavage products produced by 1 h incubation of 10 nM sense or antisense RNA 32P-labeled at the cap with 10 nM dsRNAs of the p133 series in Drosophila lysate. Length markers were generated by partial nuclease T1 digestion and partial alkaline hydrolysis (OH) of the cap-labeled target RNA. The regions targeted by the dsRNAs are indicated as black bars on both sides. The 20–23-nt spacing between the predominant cleavage sites for the 111-bp dsRNA is shown. The horizontal arrow indicates unspecific cleavage not caused by RNAi. (B) Position of the cleavage sites on sense and antisense target RNAs. The sequences of the capped 177-nt sense and 180-nt antisense target RNAs are represented in antiparallel orientation such that complementary sequence are opposing each other. The region targeted by the different dsRNAs are indicate by differently colored bars positioned between sense and antisense target sequences. Cleavage sites are indicated by circles (large circle for strong cleavage, small circle for weak cleavage). The 32P-radiolabeled phosphate group is marked by an asterisk.
Figure 3
Mapping of sense and antisense target RNA cleavage sites. (A) Denaturing gel electrophoresis of the stable 5′ cleavage products produced by 1 h incubation of 10 nM sense or antisense RNA 32P-labeled at the cap with 10 nM dsRNAs of the p133 series in Drosophila lysate. Length markers were generated by partial nuclease T1 digestion and partial alkaline hydrolysis (OH) of the cap-labeled target RNA. The regions targeted by the dsRNAs are indicated as black bars on both sides. The 20–23-nt spacing between the predominant cleavage sites for the 111-bp dsRNA is shown. The horizontal arrow indicates unspecific cleavage not caused by RNAi. (B) Position of the cleavage sites on sense and antisense target RNAs. The sequences of the capped 177-nt sense and 180-nt antisense target RNAs are represented in antiparallel orientation such that complementary sequence are opposing each other. The region targeted by the different dsRNAs are indicate by differently colored bars positioned between sense and antisense target sequences. Cleavage sites are indicated by circles (large circle for strong cleavage, small circle for weak cleavage). The 32P-radiolabeled phosphate group is marked by an asterisk.
Figure 4
21- and 22-nt RNA fragments are generated by an RNase III–like mechanism. (A) Sequences of ∼21-nt RNAs after dsRNA processing. The ∼21-nt RNA fragments generated by dsRNA processing were directionally cloned and sequenced. Oligoribonucleotides originating from the sense strand of the dsRNA are indicated as blue lines; those originating from the antisense strand are red lines. Thick bars are used if the same sequence was present in multiple clones, the number at the right indicating the frequency. The target RNA cleavage sites mediated by the dsRNA are indicated as orange circles: large circle for strong cleavage, small circle for weak cleavage (see Fig. 3B). Circles on top of the sense strand indicated cleavage sites within the sense target, and circles at the bottom of the dsRNA indicate cleavage site in the antisense target. Up to five additional nucleotides were identified in ∼21-nt fragments derived from the 3′ ends of the dsRNA. These nucleotides are random combinations of predominantly C, G, or A residues and were most likely added in an untemplated fashion during T7 transcription of the dsRNA-constituting strands. (B) Two-dimensional TLC analysis of the nucleotide composition of ∼21-nt RNAs. The ∼21-nt RNAs were generated by incubation of internally radiolabeled 504-bp _Pp_-luc dsRNA in Drosophila lysate, gel purified, and then digested to mononucleotides with nuclease P1 (top row) or ribonuclease T2 (bottom row). The dsRNA was internally radiolabeled by transcription in the presence of one of the indicated α-32P nucleoside triphosphates. Radioactivity was detected by phosphorimaging. Nucleoside 5′-monophosphates, nucleoside 3′-monophosphates, nucleoside 5′,3′-diphosphates, and inorganic phosphate are indicated as pN, Np, pNp, and pi, respectively. Black circles indicate UV-absorbing spots from nonradioactive carrier nucleotides. The 3′,5′-diphosphates (red circles) were identified by comigration with radiolabeled standards prepared by 5′-phosphorylation of nucleoside 3′-monophosphates with T4 polynucleotide kinase and γ-32P-ATP (data not shown).
Figure 4
21- and 22-nt RNA fragments are generated by an RNase III–like mechanism. (A) Sequences of ∼21-nt RNAs after dsRNA processing. The ∼21-nt RNA fragments generated by dsRNA processing were directionally cloned and sequenced. Oligoribonucleotides originating from the sense strand of the dsRNA are indicated as blue lines; those originating from the antisense strand are red lines. Thick bars are used if the same sequence was present in multiple clones, the number at the right indicating the frequency. The target RNA cleavage sites mediated by the dsRNA are indicated as orange circles: large circle for strong cleavage, small circle for weak cleavage (see Fig. 3B). Circles on top of the sense strand indicated cleavage sites within the sense target, and circles at the bottom of the dsRNA indicate cleavage site in the antisense target. Up to five additional nucleotides were identified in ∼21-nt fragments derived from the 3′ ends of the dsRNA. These nucleotides are random combinations of predominantly C, G, or A residues and were most likely added in an untemplated fashion during T7 transcription of the dsRNA-constituting strands. (B) Two-dimensional TLC analysis of the nucleotide composition of ∼21-nt RNAs. The ∼21-nt RNAs were generated by incubation of internally radiolabeled 504-bp _Pp_-luc dsRNA in Drosophila lysate, gel purified, and then digested to mononucleotides with nuclease P1 (top row) or ribonuclease T2 (bottom row). The dsRNA was internally radiolabeled by transcription in the presence of one of the indicated α-32P nucleoside triphosphates. Radioactivity was detected by phosphorimaging. Nucleoside 5′-monophosphates, nucleoside 3′-monophosphates, nucleoside 5′,3′-diphosphates, and inorganic phosphate are indicated as pN, Np, pNp, and pi, respectively. Black circles indicate UV-absorbing spots from nonradioactive carrier nucleotides. The 3′,5′-diphosphates (red circles) were identified by comigration with radiolabeled standards prepared by 5′-phosphorylation of nucleoside 3′-monophosphates with T4 polynucleotide kinase and γ-32P-ATP (data not shown).
Figure 5
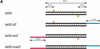
Synthetic 21- and 22-nt RNAs mediate target RNA cleavage. (A) Graphic representation of control 52-bp dsRNA and synthetic 21- and 22-nt dsRNAs. The sense strand of 21- and 22-nt short interfering RNAs (siRNAs) is shown in blue, the antisense strand in red. The sequences of the siRNAs were derived from the cloned fragments of 52- and 111-bp dsRNAs (Fig. 4A), except for the 22-nt antisense strand of duplex 5. The siRNAs in duplexex 6 and 7 were unique to the 111-bp dsRNA-processing reaction. The two 3′ overhanging nucleotides indicated in green are present in the sequence of the synthetic antisense strand of duplexes 1 and 3. Both strands of the control 52-bp dsRNA were prepared by in vitro transcription, and a fraction of transcripts may contain untemplated 3′ nucleotide addition. The target RNA cleavage sites directed by the siRNA duplexes are indicated as orange circles (see legend to Fig. 4A) and were determined as shown in Figure 5B. (B) RNA interference assay. To evaluate the efficiency of target RNA degradation, control 52-bp dsRNA (5 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) targeting full-length _Pp-_luc mRNA were tested in the translation-based RNAi assay as described in Figure 1B. The relative luminescence of target to control luciferase normalized to a buffer control (buf) is blotted; error bars indicate standard deviations calculated from at least two independent experiments. (C) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. Control 52-bp dsRNA (10 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) were incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The cleavage sites are indicated in Figure 5A. The region targeted by the 52-bp dsRNA or the sense (s) or antisense (as) strands are indicated by the black bars to the side of the gel. The cleavage sites are all located within the region of identity of the dsRNAs. For precise determination of the cleavage sites of the antisense strand, a lower percentage gel was used (data not shown).
Figure 5
Synthetic 21- and 22-nt RNAs mediate target RNA cleavage. (A) Graphic representation of control 52-bp dsRNA and synthetic 21- and 22-nt dsRNAs. The sense strand of 21- and 22-nt short interfering RNAs (siRNAs) is shown in blue, the antisense strand in red. The sequences of the siRNAs were derived from the cloned fragments of 52- and 111-bp dsRNAs (Fig. 4A), except for the 22-nt antisense strand of duplex 5. The siRNAs in duplexex 6 and 7 were unique to the 111-bp dsRNA-processing reaction. The two 3′ overhanging nucleotides indicated in green are present in the sequence of the synthetic antisense strand of duplexes 1 and 3. Both strands of the control 52-bp dsRNA were prepared by in vitro transcription, and a fraction of transcripts may contain untemplated 3′ nucleotide addition. The target RNA cleavage sites directed by the siRNA duplexes are indicated as orange circles (see legend to Fig. 4A) and were determined as shown in Figure 5B. (B) RNA interference assay. To evaluate the efficiency of target RNA degradation, control 52-bp dsRNA (5 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) targeting full-length _Pp-_luc mRNA were tested in the translation-based RNAi assay as described in Figure 1B. The relative luminescence of target to control luciferase normalized to a buffer control (buf) is blotted; error bars indicate standard deviations calculated from at least two independent experiments. (C) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. Control 52-bp dsRNA (10 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) were incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The cleavage sites are indicated in Figure 5A. The region targeted by the 52-bp dsRNA or the sense (s) or antisense (as) strands are indicated by the black bars to the side of the gel. The cleavage sites are all located within the region of identity of the dsRNAs. For precise determination of the cleavage sites of the antisense strand, a lower percentage gel was used (data not shown).
Figure 5
Synthetic 21- and 22-nt RNAs mediate target RNA cleavage. (A) Graphic representation of control 52-bp dsRNA and synthetic 21- and 22-nt dsRNAs. The sense strand of 21- and 22-nt short interfering RNAs (siRNAs) is shown in blue, the antisense strand in red. The sequences of the siRNAs were derived from the cloned fragments of 52- and 111-bp dsRNAs (Fig. 4A), except for the 22-nt antisense strand of duplex 5. The siRNAs in duplexex 6 and 7 were unique to the 111-bp dsRNA-processing reaction. The two 3′ overhanging nucleotides indicated in green are present in the sequence of the synthetic antisense strand of duplexes 1 and 3. Both strands of the control 52-bp dsRNA were prepared by in vitro transcription, and a fraction of transcripts may contain untemplated 3′ nucleotide addition. The target RNA cleavage sites directed by the siRNA duplexes are indicated as orange circles (see legend to Fig. 4A) and were determined as shown in Figure 5B. (B) RNA interference assay. To evaluate the efficiency of target RNA degradation, control 52-bp dsRNA (5 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) targeting full-length _Pp-_luc mRNA were tested in the translation-based RNAi assay as described in Figure 1B. The relative luminescence of target to control luciferase normalized to a buffer control (buf) is blotted; error bars indicate standard deviations calculated from at least two independent experiments. (C) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. Control 52-bp dsRNA (10 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) were incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The cleavage sites are indicated in Figure 5A. The region targeted by the 52-bp dsRNA or the sense (s) or antisense (as) strands are indicated by the black bars to the side of the gel. The cleavage sites are all located within the region of identity of the dsRNAs. For precise determination of the cleavage sites of the antisense strand, a lower percentage gel was used (data not shown).
Figure 6
Long 3′ overhangs on short dsRNAs inhibit RNAi. (A) Graphic representation of 52-bp dsRNA constructs. The 3′ extensions of sense and antisense strand are indicated in blue and red, respectively. The observed cleavage sites on the target RNAs are represented as orange circles analogous to Figure 4A and were determined as shown in B. (B) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. DsRNA (10 nM) was incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The major cleavage sites are indicated with a horizontal arrow and are also represented in A. The region targeted by the 52-bp dsRNA is represented as a black bar at both sides of the gel. (C) Processing of 52-bp dsRNAs with different 3′ extensions. Internally 32P-labeled dsRNAs (5 nM) were incubated in Drosophila lysate, and reaction aliquots were analyzed at the indicated time points. An RNA size marker (M) has been loaded in the left lane, and the fragment sizes are indicated. Double bands at time zero are caused by incompletely denatured dsRNA.
Figure 6
Long 3′ overhangs on short dsRNAs inhibit RNAi. (A) Graphic representation of 52-bp dsRNA constructs. The 3′ extensions of sense and antisense strand are indicated in blue and red, respectively. The observed cleavage sites on the target RNAs are represented as orange circles analogous to Figure 4A and were determined as shown in B. (B) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. DsRNA (10 nM) was incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The major cleavage sites are indicated with a horizontal arrow and are also represented in A. The region targeted by the 52-bp dsRNA is represented as a black bar at both sides of the gel. (C) Processing of 52-bp dsRNAs with different 3′ extensions. Internally 32P-labeled dsRNAs (5 nM) were incubated in Drosophila lysate, and reaction aliquots were analyzed at the indicated time points. An RNA size marker (M) has been loaded in the left lane, and the fragment sizes are indicated. Double bands at time zero are caused by incompletely denatured dsRNA.
Figure 7
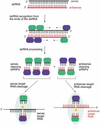
Proposed model for RNAi. RNAi is predicted to begin with processing of dsRNA (sense strand in black, antisense strand in red) to predominantly 21- and 22-nt short interfering RNAs (siRNAs). Short overhanging 3′ nucleotides, if present on the dsRNA, may be beneficial for processing of short dsRNAs. The dsRNA-processing proteins, which remain to be characterized, are represented as green and blue ovals and assemble on the dsRNA in asymmetric fashion. In our model, this is illustrated by binding of a hypothetical blue protein or protein domain with the siRNA strand in 3′ to 5′ direction while the hypothetical green protein or protein domain is always bound to the opposing siRNA strand. These proteins or a subset remain associated with the siRNA duplex and preserve its orientation, as determined by the direction of the dsRNA processing reaction. Only the siRNA sequence associated with the blue protein is able to guide target RNA cleavage. The endonuclease complex is referred to as small interfering ribonucleoprotein complex or siRNP. It is presumed here that the endonuclease that cleaves the dsRNA may also cleave the target RNA, probably by temporarily displacing the passive siRNA strand not used for target recognition. The target RNA is then cleaved in the center of the region recognized by the sequence-complementary guide siRNA. Because the cleavage site is displaced by 10–12 nt relative to the dsRNA processing site, a conformational rearrangement or a change in the composition of an siRNP must occur before target RNA cleavage.
Similar articles
- Development. Dicing up RNAs.
Ambros V. Ambros V. Science. 2001 Aug 3;293(5531):811-3. doi: 10.1126/science.1064400. Science. 2001. PMID: 11486075 No abstract available. - RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals.
Zamore PD, Tuschl T, Sharp PA, Bartel DP. Zamore PD, et al. Cell. 2000 Mar 31;101(1):25-33. doi: 10.1016/S0092-8674(00)80620-0. Cell. 2000. PMID: 10778853 - RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs.
Lipardi C, Wei Q, Paterson BM. Lipardi C, et al. Cell. 2001 Nov 2;107(3):297-307. doi: 10.1016/s0092-8674(01)00537-2. Cell. 2001. PMID: 11701121 - RNA interference: mechanisms and applications.
Fjose A, Ellingsen S, Wargelius A, Seo HC. Fjose A, et al. Biotechnol Annu Rev. 2001;7:31-57. doi: 10.1016/s1387-2656(01)07032-6. Biotechnol Annu Rev. 2001. PMID: 11686048 Review. - Antisense-RNA regulation and RNA interference.
Brantl S. Brantl S. Biochim Biophys Acta. 2002 May 3;1575(1-3):15-25. doi: 10.1016/s0167-4781(02)00280-4. Biochim Biophys Acta. 2002. PMID: 12020814 Review.
Cited by
- Transgenic tobacco lines expressing defective CMV replicase-derived dsRNA are resistant to CMV-O and CMV-Y.
Ntui VO, Kynet K, Khan RS, Ohara M, Goto Y, Watanabe M, Fukami M, Nakamura I, Mii M. Ntui VO, et al. Mol Biotechnol. 2014 Jan;56(1):50-63. doi: 10.1007/s12033-013-9681-5. Mol Biotechnol. 2014. PMID: 23820979 - Sniffing for gene-silencing efficiency of siRNAs in HeLa cells in comparison with that in HEK293T cells: correlation between knockdown efficiency and sustainability of sirnas revealed by FRET-based probing.
Shin S, Kim YS, Kim J, Kwon HM, Kim DE, Hah SS. Shin S, et al. Nucleic Acid Ther. 2013 Apr;23(2):152-9. doi: 10.1089/nat.2012.0396. Epub 2013 Feb 13. Nucleic Acid Ther. 2013. PMID: 23405948 Free PMC article. - MicroRNA-regulated viral vectors for gene therapy.
Geisler A, Fechner H. Geisler A, et al. World J Exp Med. 2016 May 20;6(2):37-54. doi: 10.5493/wjem.v6.i2.37. eCollection 2016 May 20. World J Exp Med. 2016. PMID: 27226955 Free PMC article. Review. - Therapeutic siRNA: principles, challenges, and strategies.
Gavrilov K, Saltzman WM. Gavrilov K, et al. Yale J Biol Med. 2012 Jun;85(2):187-200. Epub 2012 Jun 25. Yale J Biol Med. 2012. PMID: 22737048 Free PMC article. Review. - Conformational Dynamics of Ago-Mediated Silencing Processes.
Willkomm S, Restle T. Willkomm S, et al. Int J Mol Sci. 2015 Jul 1;16(7):14769-85. doi: 10.3390/ijms160714769. Int J Mol Sci. 2015. PMID: 26140373 Free PMC article. Review.
References
- Anandalakshmi R, Marathe R, Ge X, Herr JM, Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. - PubMed
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. - PubMed
- Bosher JM, Labouesse M. RNA interference: Genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:E31–E36. - PubMed
- Caplen NJ, Fleenor J, Fire A, Morgan RA. dsRNA-mediated gene silencing in cultured Drosophila cells: A tissue culture model for the analysis of RNA interference. Gene. 2000;252:95–105. - PubMed
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials