ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches - PubMed (original) (raw)
ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches
T Rognes. Nucleic Acids Res. 2001.
Abstract
There is a need for faster and more sensitive algorithms for sequence similarity searching in view of the rapidly increasing amounts of genomic sequence data available. Parallel processing capabilities in the form of the single instruction, multiple data (SIMD) technology are now available in common microprocessors and enable a single microprocessor to perform many operations in parallel. The ParAlign algorithm has been specifically designed to take advantage of this technology. The new algorithm initially exploits parallelism to perform a very rapid computation of the exact optimal ungapped alignment score for all diagonals in the alignment matrix. Then, a novel heuristic is employed to compute an approximate score of a gapped alignment by combining the scores of several diagonals. This approximate score is used to select the most interesting database sequences for a subsequent Smith-Waterman alignment, which is also parallelised. The resulting method represents a substantial improvement compared to existing heuristics. The sensitivity and specificity of ParAlign was found to be as good as Smith-Waterman implementations when the same method for computing the statistical significance of the matches was used. In terms of speed, only the significantly less sensitive NCBI BLAST 2 program was found to outperform the new approach. Online searches are available at http://dna.uio.no/search/
Figures
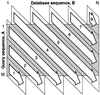
Figure 1
Computation of the diagonal scores using SIMD technology. Computation of the diagonal scores is performed efficiently in the order indicated using bands that are 32 diagonals wide.
Figure 2
Computation of the estimated gapped alignment score. The numbers within the matrix are the temporary scores (_e_i) along the diagonals from the initial computation of the diagonal scores. The numbers directly outside the matrix are the optimal ungapped alignment scores (_S_d) for each diagonal. The second numbers outside the matrix are the temporary scores (_u_d) used in the calculation of the estimated gapped alignment score (T), which in this example is 3 + 11 + 1 + 22 = 37. The BLOSUM62 matrix was used in combination with the parameters q = 11, r = 1 and c = 3 in this example. The calculations were performed in order of increasing diagonal numbers.
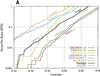
Figure 3
Comparison of database search sensitivity and selectivity. The sensitivity (coverage) versus the selectivity (EPQ) is plotted for a range of database search programs using either (A) the BLOSUM50 matrix and a 10 + 2_k_ gap penalty or (B) the BLOSUM62 matrix and a 11 + k gap penalty.
Figure 3
Comparison of database search sensitivity and selectivity. The sensitivity (coverage) versus the selectivity (EPQ) is plotted for a range of database search programs using either (A) the BLOSUM50 matrix and a 10 + 2_k_ gap penalty or (B) the BLOSUM62 matrix and a 11 + k gap penalty.
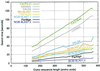
Figure 4
Comparison of database search speed. Search time versus query sequence length is plotted for the different search programs and the 11 query sequences (see Results). The search time used is the total CPU time of the fastest of three consecutive runs on a minimally loaded computer. With a database of only 29 and 128 MB of RAM, all of the database was cached in the computer’s RAM; disk reading time should then be negligible.
Similar articles
- Six-fold speed-up of Smith-Waterman sequence database searches using parallel processing on common microprocessors.
Rognes T, Seeberg E. Rognes T, et al. Bioinformatics. 2000 Aug;16(8):699-706. doi: 10.1093/bioinformatics/16.8.699. Bioinformatics. 2000. PMID: 11099256 - Accelerated Profile HMM Searches.
Eddy SR. Eddy SR. PLoS Comput Biol. 2011 Oct;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. Epub 2011 Oct 20. PLoS Comput Biol. 2011. PMID: 22039361 Free PMC article. - PARALIGN: rapid and sensitive sequence similarity searches powered by parallel computing technology.
Saebø PE, Andersen SM, Myrseth J, Laerdahl JK, Rognes T. Saebø PE, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W535-9. doi: 10.1093/nar/gki423. Nucleic Acids Res. 2005. PMID: 15980529 Free PMC article. - Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Altschul SF, et al. Nucleic Acids Res. 1997 Sep 1;25(17):3389-402. doi: 10.1093/nar/25.17.3389. Nucleic Acids Res. 1997. PMID: 9254694 Free PMC article. Review. - Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements.
Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Schäffer AA, et al. Nucleic Acids Res. 2001 Jul 15;29(14):2994-3005. doi: 10.1093/nar/29.14.2994. Nucleic Acids Res. 2001. PMID: 11452024 Free PMC article. Review.
Cited by
- Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA.
Morland I, Rolseth V, Luna L, Rognes T, Bjørås M, Seeberg E. Morland I, et al. Nucleic Acids Res. 2002 Nov 15;30(22):4926-36. doi: 10.1093/nar/gkf618. Nucleic Acids Res. 2002. PMID: 12433996 Free PMC article. - Overexpression of the LexA-regulated tisAB RNA in E. coli inhibits SOS functions; implications for regulation of the SOS response.
Weel-Sneve R, Bjørås M, Kristiansen KI. Weel-Sneve R, et al. Nucleic Acids Res. 2008 Nov;36(19):6249-59. doi: 10.1093/nar/gkn633. Epub 2008 Oct 1. Nucleic Acids Res. 2008. PMID: 18832374 Free PMC article. - PSimScan: algorithm and utility for fast protein similarity search.
Kaznadzey A, Alexandrova N, Novichkov V, Kaznadzey D. Kaznadzey A, et al. PLoS One. 2013;8(3):e58505. doi: 10.1371/journal.pone.0058505. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505522 Free PMC article. - The human ortholog of the rodent testis-specific ABC transporter Abca17 is a ubiquitously expressed pseudogene (ABCA17P) and shares a common 5' end with ABCA3.
Piehler AP, Wenzel JJ, Olstad OK, Haug KB, Kierulf P, Kaminski WE. Piehler AP, et al. BMC Mol Biol. 2006 Sep 12;7:28. doi: 10.1186/1471-2199-7-28. BMC Mol Biol. 2006. PMID: 16968533 Free PMC article. - Exploring the utility of cross-laboratory RAD-sequencing datasets for phylogenetic analysis.
Gonen S, Bishop SC, Houston RD. Gonen S, et al. BMC Res Notes. 2015 Jul 8;8:299. doi: 10.1186/s13104-015-1261-2. BMC Res Notes. 2015. PMID: 26152111 Free PMC article.
References
- Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. - PubMed
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
- Hughey R. (1996) Parallel hardware for sequence comparison and alignment. Comput. Appl. Biosci., 12, 473–479. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials