Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos - PubMed (original) (raw)
Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos
I Stancheva et al. EMBO J. 2001.
Erratum in
- Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos.
Stancheva I, Hensey C, Meehan RR. Stancheva I, et al. EMBO J. 2019 Oct 15;38(20):e103221. doi: 10.15252/embj.2019103221. EMBO J. 2019. PMID: 31612522 Free PMC article.
Abstract
DNA methylation is necessary for normal embryogenesis in animals. Here we show that loss of the maintenance methyltransferase, xDnmt1p, triggers an apoptotic response during Xenopus development, which accounts for the loss of specific cell populations in hypomethylated embryos. Hypomethylation-induced apoptosis is accompanied by a stabilization in xp53 protein levels after the mid-blastula transition. Ectopic expression of HPV-E6, which promotes xp53 degradation, prevents cell death, implying that the apoptotic signal is mediated by xp53. In addition, inhibition of caspase activation by overexpression of Bcl-2 results in the development of cellular masses that resemble embryonic blastomas. Embryonic tissue explant experiments suggest that hypomethylation alters the developmental potential of early embryo cells and that apoptosis is triggered by differentiation. Our results imply that loss of DNA methylation in differentiated somatic cells provides a signal via p53 that activates cell death pathways.
Figures
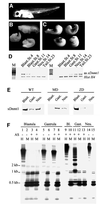
Fig. 1. Post-MBT depletion of xDnmt1 results in phenotypic abnormalities and hypomethylation. (A) Wild-type control at stage 35. (B) Antisense xDnmt1 RNA injection at the 2-cell stage depletes the maternal form of xDnmt1 (MD) and results in headless and axis-truncated phenotypes (55%), equivalent to stage 35. (C) Embryos depleted of the zygotic form of xDnmt1 RNA by antisense RNA expressed from a CMV-driven promoter (ZD) that is activated after MBT develop axis truncation (34%) and open neural tubes (30%), equivalent to stage 35. (D) RT–PCR showing the level of antisense xDnmt1 and endogenous histone H4 (Hist H4) RNAs in maternal (left set) and zygotic (right set) depleted embryos. M is a size marker; stages are indicated on top of each lane. (E) Western blot showing that xDnmt1p is absent at different stages in maternal (MD) and zygotic (ZD) _xDnmt1_-depleted embryos. (F) Southern blots of genomic DNA from MD and ZD embryos. The DNA was digested with either _Hpa_II (methylation sensitive) or _Msp_I (methylation insensitive), and probed with a 750 bp satellite I probe. This sequence is 750 bp in length, contains two _Hpa_II sites and is present in ∼2–4 × 104 copies in the genome (Lam and Carroll, 1983). In MD embryos (lanes 3 and 4), satellite I DNA is hypomethylated at blastula and remethylated at gastrula stages (lanes 7 and 8). In the ZD embryos, loss of methylation occurs later during gastrula (lanes 11 and 12) and neurula (lane 13) stages. Embryos that received the antisense RNAs are marked with +, wild type is indicated by –.
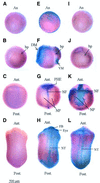
Fig. 2. Whole-mount TUNEL staining of normal and hypomethylated Xenopus embryos. Normal and methylation-deficient embryos were assayed by TUNEL for the appearance of apoptotic cells. Late blastula (A, E and I), gastrula (B, F and J), neurula (C, G and K) and tailbud (D, H and L) were assayed in WT (A–D), MD (E–H) and ZD (I–L) embryos. An MD gastrula embryo with a high number of TUNEL-positive cells, corresponding to +++ in Table I, is shown in (F). A ZD gastrula with a few TUNEL-positive cells, + in Table I, is shown in (J). A ZD neurula with the highest levels of TUNEL-positive cells, ++++, is shown in (K). Embryos in (A), (B), (C), (D) and (I) are TUNEL negative. Note that the positive staining (purple) is subsequent to, or coincident with loss of DNA methylation (Figure 1F) in MD and ZD embryos, respectively. Abbreviations: An, animal pole; bp, blastopore; DM, dorsal mesoderm; VM, ventral mesoderm; PHE, presumptive head ectoderm; NF, neural fold; NP, neural plate; FB, forebrain; NT, neural tube; Ant., anterior; and Post., posterior.
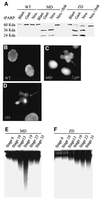
Fig. 3. Hypomethylated embryos show evidence of nuclear fragmentation and activation of caspases. (A) In vitro translated tPARP protein undergoes cleavage in extracts derived from maternal (MD) and zygotic (ZD) _xDnmt1_-depleted embryos but not in wild-type (WT) extracts. Note that xDnmt1 depletion results in tPARP cleavage during gastrula (Gast.) and neurula (Neu.) but not blastula (Blast.) stages. Note that ZD extracts have a lower caspase activity. Caspase activity can be inhibited by z-DEVD-fmk in MD and ZD neurula extracts (Neu + fmk). (B) Nuclei from wild-type neurula stage embryos stained with 4′-6-diamidine-2-phenylindole (DAPI). (C) Nuclei isolated from MD neurula stage embryos stained with DAPI. Arrow indicates a fragmented nucleus. (D) Nuclei isolated from ZD neurula stage embryos stained with DAPI. Arrow indicates a fragmented nucleus. (E) DNA isolated from MD staged embryos and separated in native agarose gels does not show fragmentation at blastula stages 7 and 10. Low molecular weight fragments appear during and after gastrulation at stages 13 and 19, and are less prominent at 23 and 35. (F) DNA from ZD embryos shows increasing fragmentation beginning at neurula stage 19 and at later stages of development (23 and 35).
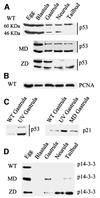
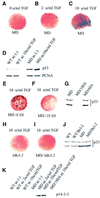
Fig. 4. Hypomethylated Xenopus embryos accumulate stable forms of p53. (A) Western blotting of staged extracts from wild-type (WT), maternally depleted xDnmt1 (MD) and post-MBT-depleted (ZD) embryos. The p53 antibody identifies two forms of xp53 at 60 and 46 kDa; note that both forms decrease as normal development proceeds. In the MD and ZD embryos the active 46 kDa form is preferentially stabilized. (B) As a loading control, all western blots were re-incubated with anti-PCNA antibodies; only the wild type is shown (WT). (C) The 46 kDa form of xp53 is stabilized in UV-treated gastrula embryos, which also leads to the induction of p21. In contrast to UV treatment, MD embryos (MD gastrula) do not induce p21 at gastrulation. (D) 14-3-3 proteins accumulate after MBT in extracts from maternally (MD) and zygotically depleted (ZD) xDnmt1 embryos but not in the wild-type (WT) embryos.
Fig. 5. The apoptotic phenotype of _xDnmt1_-deficient explants is dependent upon differentiation and can be rescued by co-injection of HPV-E6 and Bcl-2 mRNAs. (A) Animal cap explants from either antisense MD or MD/Bcl-2 co-injected blastulae are TUNEL negative in the absence of activin. An explant derived from an MD embryo is shown. (B) Animal cap explants from MD blastulae treated with a low dose of activin (2 U/ml TGF) exhibit a low level of TUNEL staining (blue). (C) High dose of activin (10 U/ml) causes death of >50% of MD animal cap explant cells as detected by the intensity of TUNEL staining (blue). (D) p53 levels are elevated compared with wild type (WT) in explants derived from antisense xDnmt1 RNA injected embryos (MD) non-induced (–) or induced (MD ac 10 U/ml TGF) with activin. A blot was reprobed for PCNA as a loading control. (E) Co-injection of a defective HPVE6 (MD/HPV-6E6) is unable to prevent hypomethylation-induced apoptosis as shown by TUNEL-positive staining (dark brown) in activin (10 U/ml TGF)-treated MD animal cap explants. (F) Co-injection of a wild-type HPVE6 (MD/HPV-18E6) is able to prevent hypomethylation-induced apoptosis in activin (10 U/ml TGF)-treated MD animal cap explants. (G) Co-injection of a wild-type HPV-E6 (MD/18E6) mRNA promotes p53 degradation in gastrula embryos whilst an N-terminal deletion of HPV-E6 (MD/6E6) does not. (H) Injection of a human Bcl-2 (hBcl-2) mRNA by itself does not lead to apoptosis in activin-treated (10 U/ml TGF) WT animal cap explants. (I) Co-injection of a human Bcl-2 mRNA (MD/hBcl-2) is able to prevent hypomethylation-induced apoptosis in activin (10 U/ml TGF)-treated MD animal caps. (J) Co-injection of a Bcl-2 mRNA does not interfere with p53 stability in wild-type (WT/Bcl-2) or maternal MD gastrula embryos. (K) 14-3-3 proteins accumulate in maternally depleted (MD), but not in the wild-type (WT) cap extracts upon activin induction. Co-injection of functional HPV-18E6 (MD/18E6 ac 10 U/ml TGF) prevents the accumulation of 14-3-3 in activin-treated MD animal cap explants. All the explants were collected 6 h after treatment and TUNEL stained for apoptosis or used for preparation of extracts.
Fig. 6. Developmental competence of _xDnmt1_-depleted ectodermal explants. The expression of specific transcripts was monitored by RT–PCR analysis of untreated and treated explants derived from wild-type embryos (WT) and embryos co-injected with antisense xDnmt1 RNA and Bcl-2 sense RNA (MD/Bcl-2). Note that the pattern of expression for differentiation-specific markers is disrupted in the _xDnmt1_-depleted explants in the absence (–) and presence (+) of activin. Histone H4 (HistH4) serves as a control as its levels do not change in response to xDnmt1 depletion or activin (+) treatment. Developmental competence was monitored by expression of Xbra, Goosecoid, muscle-specific actin (M.actin), Xnot, NCAM and epidermal keratin (Ep.ker.). A further control shows the expression of these markers in normal (WT) and maternal _xDnmt1_-depleted (MD) gastrula embryos. Note the expression of differentiation-specific markers is also disrupted in the _xDnmt1_-depleted gastrula embryos.
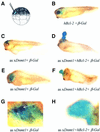
Fig. 7. Ectopic hypomethylation in cells rescued from apoptosis results in embryonic blastomas. (A) Scheme for single-cell injection into a dorsal blastomere of 32- to 64-cell embryos. (B) _β_-galactosidase and Bcl-2 sense RNAs were co-injected into a single dorsal animal blastomere of a 32- to 64-cell blastula (as indicated on the drawing). The injected embryos were allowed to develop until stage 35 and assayed for β-galactosidase activity. Note the normal appearance of the embryos and that the eye stains blue. (C) Antisense xDnmt1 RNA and _β_-galactosidase sense RNA were co-injected into a single dorsal animal blastomere of a 32- to 64-cell blastula. When assayed for β-galactosidase activity at stage 35 this resulted in staining on the forehead of the embryos amidst patches of dead cells that colocalize with β-galactosidase staining (blue). (D) Antisense xDnmt1 RNA, Bcl-2 and _β_-galactosidase sense RNA were co-injected into a single dorsal animal blastomere of a 32- to 64-cell blastula, which was allowed to develop until stage 35 and assayed for β-galactosidase activity. This resulted in the appearance of a large outgrowth of cells that grew out of the forehead and were β-galactosidase positive (blue). (E) Antisense xDnmt1 RNA and _β_-galactosidase sense RNA were co-injected into a single dorsal animal blastomere of a 32- to 64-cell blastula. When assayed for β-galactosidase activity at stage 35 this resulted in the absence of an eye amidst patches of dead cells (see G) that colocalize with β-galactosidase staining (blue). (F) Antisense xDnmt1 RNA, Bcl-2 and _β_-galactosidase sense RNA were co-injected into a single dorsal animal blastomere of a 32- to 64-cell blastula, which was allowed to develop until stage 35 and assayed for β-galactosidase activity. This resulted in the appearance of a large outgrowth of cells that grew in place of the eye and were β-galactosidase positive (blue). (G) The embryo in (E) was also assayed for apoptosis by TUNEL staining. A region corresponding to the β-galactosidase staining (blue) is shown under higher magnification to show the presence of TUNEL-positive cells (purple) highlighted by arrows. (H) The embryo in (F) was also assayed for apoptosis by TUNEL staining. A region corresponding to the β-galactosidase staining (blue) is shown under higher magnification to show the absence of TUNEL-positive cells (purple).
Similar articles
- xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function.
Dunican DS, Ruzov A, Hackett JA, Meehan RR. Dunican DS, et al. Development. 2008 Apr;135(7):1295-302. doi: 10.1242/dev.016402. Epub 2008 Feb 27. Development. 2008. PMID: 18305009 - DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos.
Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. Stancheva I, et al. Dev Biol. 2002 Mar 1;243(1):155-65. doi: 10.1006/dbio.2001.0560. Dev Biol. 2002. PMID: 11846484 - MBD4 and MLH1 are required for apoptotic induction in xDNMT1-depleted embryos.
Ruzov A, Shorning B, Mortusewicz O, Dunican DS, Leonhardt H, Meehan RR. Ruzov A, et al. Development. 2009 Jul;136(13):2277-86. doi: 10.1242/dev.032227. Development. 2009. PMID: 19502488 Free PMC article. - Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos.
Stancheva I, Meehan RR. Stancheva I, et al. Genes Dev. 2000 Feb 1;14(3):313-27. Genes Dev. 2000. PMID: 10673503 Free PMC article. - Mesoderm induction by fibroblast growth factor in early Xenopus development.
Slack JM, Darlington BG, Gillespie LL, Godsave SF, Isaacs HV, Paterno GD. Slack JM, et al. Philos Trans R Soc Lond B Biol Sci. 1990 Mar 12;327(1239):75-84. doi: 10.1098/rstb.1990.0044. Philos Trans R Soc Lond B Biol Sci. 1990. PMID: 1969663 Review.
Cited by
- Nutrition in early life, and risk of cancer and metabolic disease: alternative endings in an epigenetic tale?
Burdge GC, Lillycrop KA, Jackson AA. Burdge GC, et al. Br J Nutr. 2009 Mar;101(5):619-30. doi: 10.1017/S0007114508145883. Epub 2008 Dec 12. Br J Nutr. 2009. PMID: 19079817 Free PMC article. Review. - The role of methylation of DNA in environmental adaptation.
Flores KB, Wolschin F, Amdam GV. Flores KB, et al. Integr Comp Biol. 2013 Aug;53(2):359-72. doi: 10.1093/icb/ict019. Epub 2013 Apr 25. Integr Comp Biol. 2013. PMID: 23620251 Free PMC article. - Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development.
Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA. Rai K, et al. Mol Cell Biol. 2006 Oct;26(19):7077-85. doi: 10.1128/MCB.00312-06. Mol Cell Biol. 2006. PMID: 16980612 Free PMC article. - Chromatin-linked determinants of zygotic genome activation.
Østrup O, Andersen IS, Collas P. Østrup O, et al. Cell Mol Life Sci. 2013 Apr;70(8):1425-37. doi: 10.1007/s00018-012-1143-x. Epub 2012 Sep 11. Cell Mol Life Sci. 2013. PMID: 22965566 Free PMC article. Review. - Gene expression in Pre-MBT embryos and activation of maternally-inherited program of apoptosis to be executed at around MBT as a fail-safe mechanism in Xenopus early embryogenesis.
Shiokawa K, Aso M, Kondo T, Uchiyama H, Kuroyanagi S, Takai J, Takahashi S, Kajitani M, Kaito C, Sekimizu K, Takayama E, Igarashi K, Hara H. Shiokawa K, et al. Gene Regul Syst Bio. 2008 May 29;2:213-31. doi: 10.4137/grsb.s579. Gene Regul Syst Bio. 2008. PMID: 19787085 Free PMC article.
References
- Baylin S.B. and Herman,J.G. (2000) DNA hyper-methylation in tumorigenesis: epigenetics joins genetics. Trends Genet., 16, 168–174. - PubMed
- Bessard A.C., Garay,E., Lacronique,V., Legros,Y., Demarquay,C., Houque,A., Portefaix,J.M., Granier,C. and Soussi,T. (1998) Regulation of the specific DNA binding activity of Xenopus laevis p53: evidence for conserved regulation through the carboxy-terminus of the protein. Oncogene, 16, 883–890. - PubMed
- Chan T.A., Hermeking,H., Lengauer,C., Kinzler,K.W. and Vogelstein,B. (1999) 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature, 401, 616–620. - PubMed
- Chao D.T. and Korsmeyer,S.J. (1998) BCL-2 family: regulators of cell death. Annu. Rev. Immunol., 16, 395–419. - PubMed
- Chen R.Z., Pettersson,U., Beard,C., Jackson-Grusby,L. and Jaenisch,R. (1998) DNA hypomethylation leads to elevated mutation rates. Nature, 395, 89–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous