Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia - PubMed (original) (raw)
Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia
F Magdinier et al. Proc Natl Acad Sci U S A. 2001.
Abstract
DNA methylation of tumor suppressor genes is a common feature of human cancer. The cyclin-dependent kinase inhibitor gene p16/Ink4A is hypermethylated in a wide range of malignant tissues and the p14/ARF gene located 20 kb upstream on chromosome 9p21 is also methylated in carcinomas. p14/ARF (ARF, alternative reading frame) does not inhibit the activities of cyclins or cyclin-dependent kinase complexes; however, the importance of the two gene products in the etiology of cancer resides in their involvement in two major cell cycle regulatory pathways: p53 and the retinoblastoma protein, Rb, respectively. Distinct first exons driven from separate promoters are spliced onto the common exons 2 and 3 and the resulting proteins are translated in different reading frames. Both genes are expressed in normal cells but can be alternatively or coordinately silenced when their CpG islands are hypermethylated. Herein, we examined the presence of methyl-CpG binding proteins associated with aberrantly methylated promoters, the distribution of acetylated histones H3 and H4 by chromatin immunoprecipitation assays, and the effect of chemical treatment with 5-aza-2'-deoxycytidine (5aza-dC) and trichostatin A on gene induction in colon cell lines by quantitative reverse transcriptase-PCR. We observed that the methyl-CpG binding protein MBD2 is targeted to methylated regulatory regions and excludes the acetylated histones H3 and H4, resulting in a localized inactive chromatin configuration. When methylated, the genes can be induced by 5aza-dC but the combined action of 5aza-dC and trichostatin A results in robust gene expression. Thus, methyl-CpG binding proteins and histone deacetylases appear to cooperate in vivo, with a dominant effect of DNA methylation toward histone acetylation, and repress expression of tumor suppressor genes hypermethylated in cancers.
Figures
Figure 1
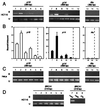
Sensitivity of PCR amplification on sonicated DNA after DNA–protein cross-linking. (A) Schematic representation of the_p15/p14/p16_ locus on human chromosome 9p21. Shaded boxes, coding exons for p15/CDKN2B/INK4b (exons 1 and 2); solid boxes, coding exons for p14/ARF (exon 1β) and p16/CDKN2A/INK4a (exons 1α, 2, and 3); arrows, transcription initiation sites. The p16 and_p14_ promoters are separated by 20 kb and both are CpG sites. Distribution of CpG sites are represented by upward strokes for_p16_ (B) and p14 (C). Lines, regions amplified by PCR from the initiation of transcription of p16 (exon 1α) (B) or p14 (exon 1β) (C). Sensitivity of the PCR amplification was monitored for ChIP experiments in which the yield of PCR product depended on the amount of input DNA and 10% of the PCR mixture (total volume, 50 μl) from serial dilution of input DNA from HCT15 colon cells was analyzed on ethidium bromide-stained 2% agarose gels. The intensity of the bands of PCR products was measured by densitometry and plotted against the amount of input DNA (from 1 to 100 ng) for each set of primers. (B) The 362-bp band corresponds to the expected size of the PCR product for the_Alu_ element (15 CpG sites, positions −1,491 to −1,132); the 395-bp band corresponds to the promoter of_p16_ (21 CpG sites, positions −494 to −101). (C) The 508-bp band corresponds to the expected size of the PCR product for the p14 promoter (43 CpG sites, positions −770 to −263).
Figure 2
ChIP analysis of the occupancy of p16 and_p14_ promoters by methyl-CpG binding proteins. (A) A quantitative analysis of PCR products was performed on chromatin immunoprecipitated with various amounts of antibodies against MBD2 and MeCP2 in HCT 15 cells. For the different conditions, the bound (B) and unbound (U) fractions were amplified with primers specific for p16, p14, and_Alu_. (B) Histograms of the ratio of the bound DNA fraction vs. the unbound DNA fraction normalized to the no antibody value (lane 6, p16; lane 12,p14; lane 15, Alu). Data are the mean ± SD for at least three experiments. (C) As a control experiment, amplification of the bound and unbound fractions was performed in HeLa where p16 and p14 are unmethylated. (D) DNA corresponding to the transcription start site of the BRCA1 gene was amplified with primers for exon 1a (16), after digestion of genomic DNA with_Hpa_II or _Cfo_I (lanes 8 and 9), or after ChIP, on input (lane 7), bound (lanes 1–6, Upper) and unbound (lanes 1–6, Lower) fractions.
Figure 3
Expression of p16 and p14 and ChIP analysis of MBD2 and acetylated histones H3 and H4 in colon cancer cell lines and HeLa. (A) p16 transcripts were coamplified by RT-PCR, with the GAPDH transcripts as an internal control; 1 μg of total RNA from HCT116, SW48, HCT15, and HeLa cells was used. PCR products (5 μl) were separated by electrophoresis on a 2% agarose gel, and positions of the bands for_p16_ (250 bp) and GAPDH (308 bp) are indicated on the left. (B) Expression of_p14_ was analyzed by RT-PCR after coamplification of_p14_ transcript (235 bp) with GAPDH (308 bp). (C) PCR analysis of DNA in chromatin immunoprecipitated with anti-MBD2 and anti-acetylated histone H3 and H4 for p16 (395 bp), p14 (508 bp), and_Alu_ (362 bp). Experiments were performed on the following cell lines: HCT116 with one allele methylated (M+) for p16 and p14 M−; SW48 with p16 M+ and_p14_ M−; HCT15 with p16 M+, p14 M+, and HeLa cells with_p16_ and p14 M−. (D) Statistical analysis (mean ± SD of at least three experiments) of the enrichment of the bound DNA fraction compared with the unbound DNA fraction for p16 and_p14,_ for each antibody and in each cell line.
Figure 4
Changes in p16 and p14 promoter occupancy and gene expression after drug treatments. (A) For each cell line, p16 expression was monitored by RT-PCR after coamplification with the GAPDH. The position of the bands is indicated to the right. For each condition, the ratio for_p16_ vs. GAPDH bands is indicated. (B) ChIP was performed with antibodies for MBD2 or acetylated histones H3 and H4, as indicated, in HCT116, SW48, HCT15, and HeLa cells after treatment with TSA, 5aza-dC, or a combination of both. Bound DNA (B) and unbound DNA (U) fractions were amplified by PCR with primers for the p16 promoter. Ratios (B/U) are below the gel. (C) For each conditions,p14 expression was monitored by RT-PCR (235 bp) after coamplification with the GAPDH (308 bp). The ratio for_p14_ expression vs. GAPDH is indicated below the gel for each cell line. (D) Bound and unbound DNA–chromatin samples immunoprecipitated with anti-MBD2 or anti-acetylated histones H3 or H4 were amplified with primers for the 3′ end of the p14 CpG island (508 bp) after treatment with TSA, 5aza-dC, or both. Ratios (B/U) are indicated below the gel for each cell line.
Figure 5
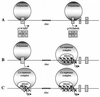
Model for the methylation-dependent silencing of p14 and_p16_ in colon cancer. (A) Normally,p14/ARF and p16/Ink4A are transcribed from different promoters and different first exons (exon 1β for_p14_ and 1α for p16) but share exons 2 and 3. The presence of methylated Alu elements between the two genes does not inhibit the elongation of transcription. ●, Methyl-CpG sites. (B) In cancer cells where p16 is hypermethylated and silenced, methyl-CpG binding proteins MBD2 (open cylinders) are recruited to the methylated CpG island containing the transcription start site and remodel the chromatin structure into an inactive state. Transcriptional silencing can be relieved by the histone deacetylase inhibitor TSA, suggesting a mechanism of gene repression that involves methyl-CpG binding proteins and a multisubunit chromatin-remodeling complex containing histone deacetylases (HDAC). The condensed region does not inhibit the expression of the p_14/ARF_ gene initiated 20 kb upstream of p16/Ink4A. (C)p14/ARF also can be methylated in colon cancer cells. MBD2 interacts with the methylated region and directs the chromatin-remodeling complex to methylated promoters. The absence of MBD2 proteins associated with the methylated Alu elements indicates that repressor complexes containing the methyl-CpG binding protein are targeted to regulatory regions, suggesting a in vivo role in the silencing of genes by DNA methylation.
Similar articles
- [Regulation of histone acetylation on the expression of cell cycle-associated genes in human colon cancer cell lines].
Chen YX, Fang JY, Lu J, Qiu DK. Chen YX, et al. Zhonghua Yi Xue Za Zhi. 2004 Feb 17;84(4):312-7. Zhonghua Yi Xue Za Zhi. 2004. PMID: 15059516 Chinese. - Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer.
Tada Y, Brena RM, Hackanson B, Morrison C, Otterson GA, Plass C. Tada Y, et al. J Natl Cancer Inst. 2006 Mar 15;98(6):396-406. doi: 10.1093/jnci/djj093. J Natl Cancer Inst. 2006. PMID: 16537832 - Increased expression of unmethylated CDKN2D by 5-aza-2'-deoxycytidine in human lung cancer cells.
Zhu WG, Dai Z, Ding H, Srinivasan K, Hall J, Duan W, Villalona-Calero MA, Plass C, Otterson GA. Zhu WG, et al. Oncogene. 2001 Nov 22;20(53):7787-96. doi: 10.1038/sj.onc.1204970. Oncogene. 2001. PMID: 11753657 - Reactivation of silenced genes and transcriptional therapy.
Chiurazzi P, Neri G. Chiurazzi P, et al. Cytogenet Genome Res. 2003;100(1-4):56-64. doi: 10.1159/000072838. Cytogenet Genome Res. 2003. PMID: 14526164 Review. - DNA methylation and gene silencing in cancer.
Baylin SB. Baylin SB. Nat Clin Pract Oncol. 2005 Dec;2 Suppl 1:S4-11. doi: 10.1038/ncponc0354. Nat Clin Pract Oncol. 2005. PMID: 16341240 Review.
Cited by
- Transcriptional regulation of hTREX84 in human cancer cells.
Guo S, Liu M, Godwin AK. Guo S, et al. PLoS One. 2012;7(8):e43610. doi: 10.1371/journal.pone.0043610. Epub 2012 Aug 27. PLoS One. 2012. PMID: 22952718 Free PMC article. - Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation.
Lorincz MC, Schübeler D, Groudine M. Lorincz MC, et al. Mol Cell Biol. 2001 Dec;21(23):7913-22. doi: 10.1128/MCB.21.23.7913-7922.2001. Mol Cell Biol. 2001. PMID: 11689684 Free PMC article. - Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins.
Carvin CD, Parr RD, Kladde MP. Carvin CD, et al. Nucleic Acids Res. 2003 Nov 15;31(22):6493-501. doi: 10.1093/nar/gkg853. Nucleic Acids Res. 2003. PMID: 14602907 Free PMC article. - Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer.
Kondo Y, Shen L, Issa JP. Kondo Y, et al. Mol Cell Biol. 2003 Jan;23(1):206-15. doi: 10.1128/MCB.23.1.206-215.2003. Mol Cell Biol. 2003. PMID: 12482974 Free PMC article. - Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice.
Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Nandakumar V, et al. Carcinogenesis. 2011 Apr;32(4):597-604. doi: 10.1093/carcin/bgq282. Epub 2010 Dec 24. Carcinogenesis. 2011. PMID: 21186298 Free PMC article. Retracted.
References
- Sherr C J. Science. 1996;274:1672–1677. - PubMed
- Weinberg R A. Cell. 1995;81:323–330. - PubMed
- Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, 3rd, Johnson B E, Skolnick M H. Science. 1994;264:436–440. - PubMed
- Stone S, Jiang P, Dayananth P, Tavtigian S V, Katcher H, Parry D, Peters G, Kamb A. Cancer Res. 1995;55:2988–2994. - PubMed
- Ruas M, Peters G. Biochim Biophys Acta. 1998;1378:F115–F177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous