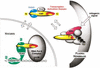TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm - PubMed (original) (raw)
TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm
A Vassilev et al. Genes Dev. 2001.
Abstract
Mammals express four highly conserved TEAD/TEF transcription factors that bind the same DNA sequence, but serve different functions during development. TEAD-2/TEF-4 protein purified from mouse cells was associated predominantly with a novel TEAD-binding domain at the amino terminus of YAP65, a powerful transcriptional coactivator. YAP65 interacted specifically with the carboxyl terminus of all four TEAD proteins. Both this interaction and sequence-specific DNA binding by TEAD were required for transcriptional activation in mouse cells. Expression of YAP in lymphocytic cells that normally do not support TEAD-dependent transcription (e.g., MPC11) resulted in up to 300-fold induction of TEAD activity. Conversely, TEAD overexpression squelched YAP activity. Therefore, the carboxy-terminal acidic activation domain in YAP is the transcriptional activation domain for TEAD transcription factors. However, whereas TEAD was concentrated in the nucleus, excess YAP65 accumulated in the cytoplasm as a complex with the cytoplasmic localization protein, 14-3-3. Because TEAD-dependent transcription was limited by YAP65, and YAP65 also binds Src/Yes protein tyrosine kinases, we propose that YAP65 regulates TEAD-dependent transcription in response to mitogenic signals.
Figures
Figure 1
Purification of TEAD-2 protein complexes from mouse cells. (A) A TEAD-2 protein complex was purified either from 3T3 cells (lanes 1–3) or from 3T3 cells expressing FH-TEAD-2 protein (lanes 4–6). FLAG tagged proteins in the extracts were bound to anti-FLAG resin, washed with buffer, and then eluted with FLAG peptide (lanes 2, 5). The FLAG eluate was then bound to an anti-HA resin, unbound proteins removed with buffer (lanes 1, 4), and the bound proteins eluted with HA peptide (lanes 3, 6). Aliquots of each fraction were subjected to SDS-PAGE, and the proteins were stained with silver. A set of standard proteins (Invitrogen) were fractionated in parallel on each gel to determine molecular weights (kD). Twelve proteins (T1-T12) were associated specifically with FH-TEAD-2 (lane 6), because they were absent under the same conditions when nonexpressing cells were used (lane 3). Three proteins were identified by mass spectroscopy as MUPP1 (T8), YAP65 (abbreviated YAP) (T1/2), and actin. TEAD-2 was identified by its reactivity with anti-FLAG and anti-TEAD-2 peptide antibodies, and by its molecular weight. (B) The HA eluate (see A, lane 6) was fractionated by sedimentation in a glycerol gradient, and individual fractions were subjected to SDS-PAGE and then stained with silver. (C) Aliquots of each fraction in B were fractionated by SDS-PAGE and subjected to immunoblotting with anti-FLAG antibody to quantify the amount of FH-TEAD-2 in each fraction.
Figure 2
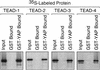
Identification of the YAP protein binding domain in TEAD-2 protein. (A) Each TEAD-2 protein used in B and C is shown, together with the map number of their terminal amino acid and the efficiency with which they bound full-length YAP protein. Each data point is the mean of 3–5 independent determinations (
sem
was 6% to 11% of the mean value). The DNA binding domain (aa 40–112) (Kaneko and DePamphilis 1998), the total YAP binding domain (aa 115–445), and the essential YAP binding domain (aa 224–445) are indicated by shaded blocks. The transcriptional activation domain mapped in human TEAD-1 (Hwang et al. 1993) is indicated by a solid bar. (B) To identify the YAP binding domain in TEAD-2 protein, full-length GST–YAP protein was attached to beads and incubated either with full-length [35S]TEAD-2 protein (TEAD–2) or with the indicated TEAD-2 deletion mutant (A to O). The amount of full-length [35S]TEAD–2 bound to full-length GST–YAP was taken as “100% YAP binding.” YAP binding was corrected for variation in the amount of [35S]TEAD-2 added to each assay ([35S]TEAD-2 input). Binding of [35S]TEAD–2 to GST alone was not detected (GST). (C) To identify fragments of TEAD-2 protein that can bind YAP, full-length GST–TEAD-2 protein or protein fragment (GST-P to S) were attached to beads and incubated with full-length [35S]YAP protein.
Figure 3
YAP binds to all four TEAD proteins. To determine whether or not full-length TEAD-1,-2, -3, and -4 proteins can bind YAP with similar efficiency, full-length GST–YAP or GST alone was attached to beads as in Figure 2 and incubated with the indicated full-length [35S]TEAD protein.
Figure 4
Identification of the TEAD protein binding domain in YAP. (A) Each YAP protein used in B is shown, together with the map number of their terminal amino acid and the efficiency with which they bound full-length TEAD–2 protein. The TEAD binding domain (aa 32–139), the 14-3-3 binding domain (Kanai et al. 2000), protein binding domains WW, SH3, and TWL (shaded bars), and the YAP transcriptional activation domain (solid bar) are indicated (Yagi et al. 1999). (B) The indicated full-length GST–YAP protein (YAP), YAP(–TEAD bd), a deletion of amino acids 77 to 96, and YAP protein fragments (A to S) were attached to beads and then incubated with full-length [35S]TEAD-2 protein to determine their binding efficiency (“% TEAD-2 binding”), as described in Figure 2.
Figure 5
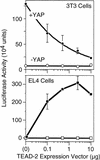
TEAD-dependent transcription requires YAP. Mouse 3T3 embryonic fibroblasts (which express YAP and TEAD-1, -2, -3, and -4 genes) or EL-4 T-lymphocytes (which express neither YAP nor TEAD proteins) were transfected with a mixture of three plasmids: pGT4Tluc, which expresses firefly luciferase only in the presence of a TEAD transcription factor; an increasing amount of pCI[H–TEAD–2], which expresses mouse H–TEAD–2 to activate transcription of the luciferase gene in pGT4Tluc; and pRI(βgal), which expresses Escherichia coli β–galactosidase to measure transfection efficiency. The amount of luciferase activity (measured as light units) was normalized to the amount of β–galactosidase activity in each assay. Where indicated (solid symbols), a fixed amount of pSI(FH-mYAP) was included to produce mouse YAP protein to determine whether or not YAP was required for TEAD-dependent transcription. The mean values (±
sem
) from three independent experiments are given.
Figure 6
TEAD-dependent transcription by all four TEAD transcription factors requires YAP with a functional TEAD binding domain and TEAD with a functional DNA binding domain. (A) Conditions for comparing TEAD-1, -2, -3, and -4 proteins in EL4 cells were the same as in Figure 5, except that each assay contained 0.1 μg pCI[H-TEAD1, -2, -3, or -4], and 10 μg pSI[YAP] or pSI when pSI[YAP] was omitted. YAP(-SH3 bd) is a mutation in the SH3 binding domain that has three proline residues at positions 267–269 mutated to glycines destroying the Src/Yes kinase SH3 domain consensus binding sequence (Sparks et al. 1996). TEAD(-DNA bd) contains a G to D amino acid substitution at position 74 in the DNA binding domain (Kaneko and DePamphilis 1998). To facilitate comparison, luciferase activity observed for each TEAD protein in the presence of YAP was taken as 100%. Luciferase activity (104 units) was 29 (TEAD-1), 29 (TEAD-2), 30 (TEAD-3) and 12 (TEAD-4). (B) Conditions for evaluating YAP activity in 3T3 embryonic fibroblasts, C2C12 muscle fibroblasts, EL4 T-lymphocytes, and MPC-11 B-lymphocytes were the same as in Figure 5, except that 0.1 μg pCI[H-TEAD-2] was used to produce H-TEAD-2, and 10 μg of either pSI[YAP] or pSI[YAP(-TEAD bd) was used to produce either YAP or a deletion mutant of YAP that failed to bind TEAD protein (Fig. 4). The mean values (±
sem
) from four independent experiments are given. Luciferase activity observed in the presence of YAP was taken as 100% for each cell line.
Figure 7
Comparison of YAP protein complexes with TEAD-2 protein complexes assembled in mouse cells. (A) FLAG-HA-tagged protein complexes were purified from 3T3 cells expressing either FH–TEAD–2 protein (lanes 1, 2) or FH-YAP protein (lanes 3, 4) by sequential affinity chromatography, as described in Figure 1, fractionated in parallel by SDS-PAGE, and then stained with silver to compare peptide bands. A duplicate of lane 4 was immunoblotted with anti-14-3-3 serum (Upstate Biotechnology). (B) Formation of a YAP/TEAD/DNA complex. Purified FH-YAP and FH–TEAD–2 complexes, or the indicated purified proteins were mixed with [32P]DNA containing either a wild-type PyV enhancer sequence ([32P]DNA control) that does not bind TEAD proteins, or a single base pair change in this PyV sequence that converts it into a GT-IIC sequence that does bind TEAD proteins ([32P]DNA TEAD binding site), and then fractionated by gel electrophoresis. The positions of [32P]DNA/TEAD complexes and [32P]DNA/TEAD/YAP complexes are indicated. All samples were run on the same gel. Some lanes were deleted for simplicity, and a lighter exposure of the two “FH–YAP complex” lanes is shown to see the presence of two bands.
Figure 8
Subcellular localization of TEAD-2 and YAP proteins. Cells in _A–_C were expressing FH-TEAD-2, whereas cells in D–F were expressing FH-YAP. Arrows in A–C identify a nucleus that expressed endogenous TEAD-2, but not FH-TEAD-2.
Figure 9
Summary of TEAD and YAP65 interactions (see Discussion).
Similar articles
- The PDZ-binding motif of Yes-associated protein is required for its co-activation of TEAD-mediated CTGF transcription and oncogenic cell transforming activity.
Shimomura T, Miyamura N, Hata S, Miura R, Hirayama J, Nishina H. Shimomura T, et al. Biochem Biophys Res Commun. 2014 Jan 17;443(3):917-23. doi: 10.1016/j.bbrc.2013.12.100. Epub 2013 Dec 28. Biochem Biophys Res Commun. 2014. PMID: 24380865 - The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members.
Mahoney WM Jr, Hong JH, Yaffe MB, Farrance IK. Mahoney WM Jr, et al. Biochem J. 2005 May 15;388(Pt 1):217-25. doi: 10.1042/BJ20041434. Biochem J. 2005. PMID: 15628970 Free PMC article. - Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level.
Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Omerovic J, et al. Exp Cell Res. 2004 Apr 1;294(2):469-79. doi: 10.1016/j.yexcr.2003.12.002. Exp Cell Res. 2004. PMID: 15023535 - Serum Inter-α-inhibitor activates the Yes tyrosine kinase and YAP/TEAD transcriptional complex in mouse embryonic stem cells.
Pijuan-Galitó S, Tamm C, Annerén C. Pijuan-Galitó S, et al. J Biol Chem. 2014 Nov 28;289(48):33492-502. doi: 10.1074/jbc.M114.580076. Epub 2014 Oct 9. J Biol Chem. 2014. PMID: 25301940 Free PMC article. - Emerging roles of TEAD transcription factors and its coactivators in cancers.
Pobbati AV, Hong W. Pobbati AV, et al. Cancer Biol Ther. 2013 May;14(5):390-8. doi: 10.4161/cbt.23788. Epub 2013 Feb 4. Cancer Biol Ther. 2013. PMID: 23380592 Free PMC article. Review.
Cited by
- Discovering the inhibition of YAP/TEAD Hippo signaling pathway via new pyrazolone derivatives: synthesis, molecular docking and biological investigations.
Noser AA, Salem MM, Baren MH, Selim AI, ElSafty EM. Noser AA, et al. Sci Rep. 2024 Nov 21;14(1):28859. doi: 10.1038/s41598-024-79992-x. Sci Rep. 2024. PMID: 39572674 Free PMC article. - A cofactor-induced repressive type of transcription factor condensation can be induced by synthetic peptides to suppress tumorigenesis.
Tang Y, Chen F, Fang G, Zhang H, Zhang Y, Zhu H, Zhang X, Han Y, Cao Z, Guo F, Wang W, Ye D, Ju J, Tan L, Li C, Zhao Y, Zhou Z, An L, Jiao S. Tang Y, et al. EMBO J. 2024 Nov;43(22):5586-5612. doi: 10.1038/s44318-024-00257-4. Epub 2024 Oct 2. EMBO J. 2024. PMID: 39358623 Free PMC article. - YAP-TEAD inhibition is associated with upregulation of an androgen receptor mediated transcription program providing therapeutic escape.
Alva-Ruiz R, Watkins RD, Tomlinson JL, Yonkus JA, Abdelrahman AM, Conboy CB, Jessen E, Werneburg NW, Kuipers H, Sample JW, Gores GJ, Ilyas SI, Truty MJ, Smoot RL. Alva-Ruiz R, et al. FEBS Open Bio. 2024 Nov;14(11):1873-1887. doi: 10.1002/2211-5463.13901. Epub 2024 Sep 19. FEBS Open Bio. 2024. PMID: 39300603 Free PMC article. - Going Rogue: Mechanisms, Regulation, and Roles of Mutationally Activated G_α_ in Human Cancer.
Dwyer MB, Aumiller JL, Wedegaertner PB. Dwyer MB, et al. Mol Pharmacol. 2024 Oct 17;106(5):198-215. doi: 10.1124/molpharm.124.000743. Mol Pharmacol. 2024. PMID: 39187387 Review.
References
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. New York: John Wiley & Sons, Inc.; 1997.
- Azakie A, Larkin SB, Farrance IK, Grenningloh G, Ordahl CP. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J Biol Chem. 1996;271:8260–8265. - PubMed
- Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275:30801–30805. - PubMed
- Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes & Dev. 1994;8:2293–2301. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous