Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen - PubMed (original) (raw)
Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen
D Tsuchimoto et al. Nucleic Acids Res. 2001.
Abstract
In human cells APE1 is the major AP endonuclease and it has been reported to have no functional mitochondrial targeting sequence (MTS). We found that APE2 protein possesses a putative MTS. When its N-terminal 15 amino acid residues were fused to the N-terminus of green fluorescent protein and transiently expressed in HeLa cells the fusion protein was localized in the mitochondria. By electron microscopic immunocytochemistry we detected authentic APE2 protein in mitochondria from HeLa cells. Western blotting of the subcellular fraction of HeLa cells revealed most of the APE2 protein to be localized in the nuclei. We found a putative proliferating cell nuclear antigen (PCNA)-binding motif in the C-terminal region of APE2 and showed this motif to be functional by immunoprecipitation and in vitro pull-down binding assays. Laser scanning immunofluorescence microscopy of HeLa cells demonstrated both APE2 and PCNA to form foci in the nucleus and also to be co-localized in some of the foci. The incubation of HeLa cells in HAT medium containing deoxyuridine significantly increased the number of foci in which both molecules were co-localized. Our results suggest that APE2 participates in both nuclear and mitochondrial BER and also that nuclear APE2 functions in the PCNA-dependent BER pathway.
Figures
Figure 1
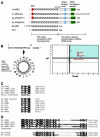
APE2 and other members of the APE2/APN2 family. (A) Structures of APE2/APN2 family proteins, E.coli XTH and human APE1 are shown. The regions conserved in the XTH family (hatched box) and the unique C-terminal region of the APE2/APN2 family are indicated. Colored boxes indicate the three unique subregions found in the APE2/APN2 family. Red, mitochondrial targeting sequence (MTS); blue, PCNA-binding motif; green, TOP3 homologous subregion. (B) Predicted MTS in APE2 protein and comparison of the MTS found in APE2/APN2 family proteins. The MitoProt II program predicted that the first 18 residues of APE2 protein form an amphipathic helix with three arginine residues but no acidic residue, as shown in the left panel. No putative cleavage site was predicted. Basic residues are shown in circles and hydrophobic residues are in boxes. Each value for µ_H_ and H_max calculated for COX8 and various repair enzymes, including the APE2/APN2 family present in mitochondria, by the MitoProt II program are plotted on the right. Red circles indicate the APE2/APN2 family proteins. Green, blue and black triangles show UNG1 (13), MYHα3 (12,43) and OGG1-2a (11), respectively. Green and blue squares show DNA polymerase γ (Polγ) (17) and DNA ligase III (LigIII) (18), respectively. An inverted triangle and a rhombus show MTH1d (accession no. BAA83791) and COX8 (31), respectively. The thick and thin lines indicate the upper and lower cut values of µ_H and _H_max, respectively. (C) Conserved PCNA-binding motifs in the APE2/APN2 family are shown and compared with those of other BER-related proteins and p21/Cip1/Waf1, which were pointed out by Warbrick (35), as well as putative motifs in OGG1 (11) and MYH (accession no. AAC50618). The first glutamine residue in the consensus is altered to an asparagine residue in putative motifs found in ScAPN2 and OGG1. In the motif consensus h indicates residues with moderately hydrophobic side chains, such as leucine, isoleucine or methionine, and a indicates residues with highly hydrophobic aromatic side chains, e.g. phenylalanine and tyrosine. (D) TOP3 homologous sequences of the APE2/APN2 family are aligned with the C-terminal repeats found in human TOP3α protein (accession no. Q13427).
Figure 2
Expression of APE2 mRNA and APE2 protein in cultured human cells and human tissues. (A) RT–PCR for APE2 mRNA. A 347 bp cDNA fragment corresponding to the APE2 coding region (nt 13–359) was amplified from total RNA prepared from HeLa and Jurkat cells and human tissues by RT–PCR. Human β-actin mRNA was amplified as an internal control. (B) Immunological detection of APE2 polypeptide in cultured human cells. Authentic and recombinant APE2 proteins were examined by western blot analysis using anti-APE2 pre-adsorbed with TrxA–APE2–Sepharose or TrxA–Sepharose. Whole cell lysate of HeLa MRV (5.0 × 105 cells in lane 1) or HeLa MR:APE2-HA (1.0 and 0.25 × 105 cells in lanes 2 and 3, respectively) were subjected to the analysis. (C) Whole cell lysate (lane W), crude isolated nuclei (lane N) and crude cytoplasmic fraction (lane C) prepared from 5.0 × 105 HeLa MRV cells were subjected to western blotting with anti-APE2.
Figure 3
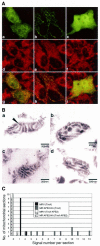
Mitochondrial localization of APE2. (A) The N-terminal 15 amino acid sequence of APE2 functions as a mitochondrial targeting sequence. Laser scanning fluorescence microscopy of cells expressing APE2–EGFPm (a, d and g), COX8–EGFPm (b, e and h) or EGFP (c, f and i) were performed after incubation with MitoTracker Red CM-H2Ros for 1 h. Signals for EGFP and MitoTracker are shown in green (a–c) and red (d–f), respectively. In merged images (g–i) the co-localized signals for EGFP and MitoTracker are shown in yellow. (B) Submitochondrial localization of human APE2 protein, determined by electron microscopic immunocytochemistry. Mitochondria were prepared from HeLa MRV (a and b) and HeLa MR:APE2-HA (c and d) cells. APE2 signals were examined by electron microscopic immunocytochemistry, using anti-APE2 pre-adsorbed to TrxA (a and c) or TrxA–APE2–Sepharose (b and d) in combination with protein A–gold. A signal for authentic APE2 is shown with an arrowhead in (a). (C) Distribution of APE2 signals in a mitochondrial section determined by electron microscopic immunocytochemistry. APE2 signals detected in each mitochondrial section as seen in (B) were counted for 10 sections in each sample prepared from HeLa MRV (solid column and open column) or HeLa MR:APE2-HA (gray and hatched columns), which were treated with anti-APE2 pre-adsorbed to TrxA–Sepharose (solid and gray columns) or TrxA–APE2–Sepharose (open and hatched columns) and the number of sections (_y_-axis) with a given number of gold signals in each section (_x_-axis) are shown in the histogram.
Figure 4
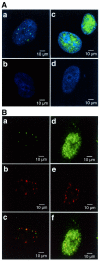
Distribution of APE2 protein in the nuclei of human cells. (A) APE2 forms nuclear foci in cultured human cells. APE2 protein in HeLa MRV (a and b) and HeLa MR:APE2-HA cells (c and d) were examined by laser scanning fluorescence microscopy, using anti-APE2 pre-adsorbed to TrxA–Sepharose (a and c) or TrxA–APE2–Sepharose (b and d), in combination with Alexa Fluor 488-labeled second antibody. Nuclei were counterstained with TOTO-3 (a–d). Alexa Fluor 488 and TOTO-3 signals are shown in green and blue, respectively. (B) Co-localization of APE2 and PCNA in nuclear foci. HeLa MRV (a–c) and HeLa MR:APE2-HA (d–f) cells were immunostained with anti-APE2/Alexa Fluor 488-labeled second antibody and anti-PCNA/Alexa Fluor 594-labeled second antibody and were subjected to laser scanning fluorescence microscopy. APE2 and PCNA signals are shown in green (a and d) and red (b and e), respectively. In merged images in (c) and (f) co-localized signals are shown in yellow.
Figure 5
Molecular interaction of APE2 and PCNA. (A) Co-immunoprecipitation of APE2 with anti-PCNA. Whole cell extracts from HeLa MRV and HeLa MR:APE2-HA cells were incubated with anti-PCNA or anti-β-galactosidase in combination with protein A–agarose for immunoprecipitation. APE2 and PCNA antigens in the immune complex were examined by western blotting with anti-APE2 (upper) and anti-PCNA (lower). (B) Interaction of recombinant PCNA immobilized on resin with APE2–HA protein expressed in HeLa MR:APE2-HA cells. His–PCNA and TrxA proteins immobilized on TALON resin were incubated with whole cell extract prepared from HeLa MR:APE2-HA cells. APE2–HA bound to each resin (2 µl bed volume with 2.6 µg His–PCNA or 2.4 µg TrxA, respectively) were detected by western blotting with anti-APE2. (C) In vitro interaction of TrxA–APE2 protein immobilized on resin and purified recombinant PCNA. TrxA–APE2 and TrxA immobilized on TALON resin (10 µl bed volume with 4.8 µg intact TrxA–APE2 or 12 µg TrxA, respectively) were incubated with 300 ng purified recombinant PCNA (rPCNA) in binding buffer containing 50, 150 or 500 mM NaCl. PCNA bound to 2 µl of each resin was subjected to western blotting. (D) Peptides with the sequence of the PCNA-binding motif found in APE2 interact with PCNA. Whole cell extracts prepared from HeLa MRV cells were incubated with synthetic peptides carrying the PCNA-binding motif in human APE2, wild-type (YF, biotin-SGSGSRGQKNLKSYFQPSPSCPQA) or mutant (AA, biotin-SGSGSRGQKNLKSAAQPSPSCPQA), and the complex was recovered with the aid of streptavidin–agarose.
Figure 6
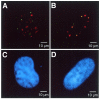
Increased misincorporation of dUMP into DNA promotes the association of APE2 and PCNA in nuclear foci. HeLa MRV cells were incubated in medium supplemented with (B and D) or without (A and C) HAT and dUrd for 6 h. The cells were immunostained with anti-APE2/Alexa Fluor 488-labeled second antibody and anti-PCNA/Alexa Fluor 594-labeled second antibody and were subjected to laser scanning fluorescence microscopy. The APE2 and PCNA signals are shown in green and red, respectively, and the co-localized signals are shown in yellow (A and B). The nuclei were counterstained with TOTO-3 (C and D).
Similar articles
- Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair.
Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Parker A, et al. J Biol Chem. 2001 Feb 23;276(8):5547-55. doi: 10.1074/jbc.M008463200. Epub 2000 Nov 22. J Biol Chem. 2001. PMID: 11092888 - Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress.
Akbari M, Otterlei M, Peña-Diaz J, Krokan HE. Akbari M, et al. Neuroscience. 2007 Apr 14;145(4):1201-12. doi: 10.1016/j.neuroscience.2006.10.010. Epub 2006 Nov 13. Neuroscience. 2007. PMID: 17101234 - Replication protein A stimulates proliferating cell nuclear antigen-dependent repair of abasic sites in DNA by human cell extracts.
Dianov GL, Jensen BR, Kenny MK, Bohr VA. Dianov GL, et al. Biochemistry. 1999 Aug 24;38(34):11021-5. doi: 10.1021/bi9908890. Biochemistry. 1999. PMID: 10460157 - Asbestos increases mammalian AP-endonuclease gene expression, protein levels, and enzyme activity in mesothelial cells.
Fung H, Kow YW, Van Houten B, Taatjes DJ, Hatahet Z, Janssen YM, Vacek P, Faux SP, Mossman BT. Fung H, et al. Cancer Res. 1998 Jan 15;58(2):189-94. Cancer Res. 1998. PMID: 9443389 - Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III.
Hadi MZ, Wilson DM 3rd. Hadi MZ, et al. Environ Mol Mutagen. 2000;36(4):312-24. Environ Mol Mutagen. 2000. PMID: 11152564
Cited by
- Base Excision Repair in the Immune System: Small DNA Lesions With Big Consequences.
Stratigopoulou M, van Dam TP, Guikema JEJ. Stratigopoulou M, et al. Front Immunol. 2020 May 29;11:1084. doi: 10.3389/fimmu.2020.01084. eCollection 2020. Front Immunol. 2020. PMID: 32547565 Free PMC article. Review. - Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1.
Li M, Zhong Z, Zhu J, Xiang D, Dai N, Cao X, Qing Y, Yang Z, Xie J, Li Z, Baugh L, Wang G, Wang D. Li M, et al. J Biol Chem. 2010 May 14;285(20):14871-14881. doi: 10.1074/jbc.M109.069591. Epub 2010 Mar 15. J Biol Chem. 2010. PMID: 20231292 Free PMC article. - Role of PCNA-dependent stimulation of 3'-phosphodiesterase and 3'-5' exonuclease activities of human Ape2 in repair of oxidative DNA damage.
Burkovics P, Hajdú I, Szukacsov V, Unk I, Haracska L. Burkovics P, et al. Nucleic Acids Res. 2009 Jul;37(13):4247-55. doi: 10.1093/nar/gkp357. Epub 2009 May 13. Nucleic Acids Res. 2009. PMID: 19443450 Free PMC article. - Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells.
Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Chattopadhyay R, et al. Nucleic Acids Res. 2006 Apr 14;34(7):2067-76. doi: 10.1093/nar/gkl177. Print 2006. Nucleic Acids Res. 2006. PMID: 16617147 Free PMC article. - Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity.
Doré AS, Kilkenny ML, Jones SA, Oliver AW, Roe SM, Bell SD, Pearl LH. Doré AS, et al. Nucleic Acids Res. 2006;34(16):4515-26. doi: 10.1093/nar/gkl623. Epub 2006 Aug 31. Nucleic Acids Res. 2006. PMID: 16945955 Free PMC article.
References
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
- Krokan H.E., Nilsen,H., Skorpen,F., Otterlei,M. and Slupphaug,G. (2000) Base excision repair of DNA in mammalian cells. FEBS Lett., 476, 73–77. - PubMed
- Lindahl T. (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res., 462, 129–135. - PubMed
- Memisoglu A. and Samson,L. (2000) Base excision repair in yeast and mammals. Mutat. Res., 451, 39–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous