Spectroscopic changes during a single turnover of biotin synthase: destruction of a [2Fe-2S] cluster accompanies sulfur insertion - PubMed (original) (raw)
Spectroscopic changes during a single turnover of biotin synthase: destruction of a [2Fe-2S] cluster accompanies sulfur insertion
N B Ugulava et al. Biochemistry. 2001.
Abstract
Biotin synthase catalyzes the insertion of a sulfur atom between the saturated C6 and C9 carbons of dethiobiotin. Catalysis requires AdoMet and flavodoxin and generates 5'-deoxyadenosine and methionine, suggesting that biotin synthase is an AdoMet-dependent radical enzyme. Biotin synthase (BioB) is aerobically purified as a dimer of 38.4 kDa monomers that contains 1-1.5 [2Fe-2S](2+) clusters per monomer and can be reconstituted with exogenous iron, sulfide, and reductants to contain up to two [4Fe-4S] clusters per monomer. The iron-sulfur clusters may play a dual role in biotin synthase: a reduced iron-sulfur cluster is probably involved in radical generation by mediating the reductive cleavage of AdoMet, while recent in vitro labeling studies suggest that an iron-sulfur cluster also serves as the immediate source of sulfur for the biotin thioether ring. Consistent with this dual role for iron-sulfur clusters in biotin synthase, we have found that the protein is stable, containing one [2Fe-2S](2+) cluster and one [4Fe-4S](2+) cluster per monomer. In the present study, we demonstrate that this mixed cluster state is essential for optimal activity. We follow changes in the Fe and S content and UV/visible and EPR spectra of the enzyme during a single turnover and conclude that during catalysis the [4Fe-4S](2+) cluster is preserved while the [2Fe-2S](2+) cluster is destroyed. We propose a mechanism for incorporation of sulfur into dethiobiotin in which a sulfur atom is oxidatively extracted from the [2Fe-2S](2+) cluster.
Figures
Scheme 1:
Conversion of Dethiobiotin to Biotin Catalyzed by Biotin Synthase
Figure 1:
Formation of biotin catalyzed by BioB at 37 °C (circles and solid curve) and 25 °C (squares and dashed curve). BioB (35 μM monomer) was preincubated with AdoMet, flavodoxin, flavodoxin reductase, NADPH, FeCl3, Na2S, and DTT in 50 mM Tris-HCl and 10 mM KCl, pH 8.0, under argon and the reaction initiated by addition of dethiobiotin. Samples (100 μL) were withdrawn at various intervals, quenched with 5 μL of saturated sodium acetate, pH 4, and analyzed by HPLC as described in Materials and Methods.
Figure 2:
UV/visible spectrum of BioB containing both [2Fe-2S]2+ and [4Fe-4S]2+ clusters (∼80 μM, solid curve). BioB was incubated under argon with FeCl3, Na2S, DTT, flavodoxin, flavodoxin reductase, and NADPH and then reisolated by anaerobic gel filtration chromatography. The dashed curves show BioB (100 μM) with ∼1.4 [2Fe-2S]2+ clusters per monomer [dashed curve, λmax at 452 nm (24)] and reductively reconstituted (20) with ∼1 [4Fe-4S]2+ cluster (dotted curve, λmax at 410 nm).
Figure 3:
Formation of biotin by BioB initially containing 1:1 [4Fe-4S]2+ and [2Fe-2S]2+ clusters (solid circles), two [4Fe-4S]2+ clusters (solid squares), two [4Fe-4S]+ clusters (open circles), and 1.4 [2Fe-2S]2+ clusters (triangles). Enzyme was prepared by chemical reduction and reconstitution as described in Materials and Methods and repurified by anaerobic gel filtration chromatography. Assays were performed as described in Materials and Methods, except that FeCl3, Na2S, and DTT were not added. Samples were withdrawn at intervals and quenched in saturated sodium acetate, pH 4, and biotin was quantified by HPLC analysis.
Figure 4:
Changes in the UV/visible spectrum of BioB observed during a single turnover. BioB (∼35 μM) was generated with 1:1 [2Fe-2S]2+ and [4Fe-4S]2+ clusters and was preincubated with AdoMet (500 μM), flavodoxin (10 μM), flavodoxin reductase (4 μM), and NADPH (1 mM) in an anaerobic cuvette, and the reaction was initiated by addition of dethiobiotin (200 μM) at 23 °C. UV/visible spectra were recorded at intervals (10, 30, 60, and 90 min shown) and show a gradual decrease in absorbance from 400 to 600 nm. The broad absorbance from 550 to 700 nm is due to the presence of flavodoxin semiquinone. (Inset) The difference spectrum (90 min - 1 min) shows a maximal decrease in absorbance at 460 nm, consistent with the loss of a [2Fe-2S]2+ cluster.
Figure 5:
The production of biotin correlates with the decrease in absorbance at 460 nm. Biotin formation (squares) and the decrease in absorbance at 460 nm (circles) are both fit with a single rate constant of 0.03 min-1 at 23 °C. Assay conditions are as described for Figure 4. The absorbance at 460 nm was monitored, samples were withdrawn at intervals and quenched in saturated sodium acetate, pH 4, and biotin was quantified by HPLC analysis. The rate of biotin production is decreased relative to Figure 3 due to the decreased temperature.
Figure 6:
(A) EPR spectrum of BioB prior to and during a single turnover of biotin production. BioB (100 μM monomer) was generated with 1:1 [2Fe-2S]2+ and [4Fe-4S]2+ clusters and was preincubated with excess substrates in the presence or absence of FeCl3, Na2S, and DTT, and samples (∼200 μL) were withdrawn for EPR analysis. The top curves show that no signal is observed in the absence of enzyme or dethiobiotin. In the presence of FeCl3, Na2S, and DTT the time-dependent formation of a new EPR-active species is observed. This species is not observed in the absence of FeCl3, Na2S, and DTT. Biotin (∼0.9 equiv per monomer) is formed under both conditions. (B) EPR spectrum of BioB after a single turnover and following chemical reduction. BioB with 1:1 [2Fe-2S]2+ and [4Fe-4S]2+ clusters was incubated with the substrates FeCl3, Na2S, and DTT for 120 min and then repurified by anaerobic gel filtration chromatography, resulting in ca. 2-fold dilution. The EPR spectrum is otherwise identical to the final spectrum in panel A. The protein was then reduced by incubation with dithionite (2 mM) for 30 min. The final reduced spectrum resembles the EPR spectrum previously reported for reductively reconstituted [4Fe-4S]+ enzyme (20), shown as a dashed curve scaled for comparison. The small resonance at g = 2.05 is not observed in all enzyme preparations and may be due to a contaminating protein or metal.
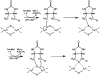
Scheme 2:
Proposed Mechanism for the Oxidative Insertion of Sulfide into Dethiobiotin from the [2Fe-2S]2+ Cluster in BioB
Similar articles
- Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions.
Ugulava NB, Gibney BR, Jarrett JT. Ugulava NB, et al. Biochemistry. 2001 Jul 27;40(28):8343-51. doi: 10.1021/bi0104625. Biochemistry. 2001. PMID: 11444981 Free PMC article. - Iron-sulfur cluster interconversions in biotin synthase: dissociation and reassociation of iron during conversion of [2Fe-2S] to [4Fe-4S] clusters.
Ugulava NB, Gibney BR, Jarrett JT. Ugulava NB, et al. Biochemistry. 2000 May 2;39(17):5206-14. doi: 10.1021/bi9926227. Biochemistry. 2000. PMID: 10819988 Free PMC article. - Role of the [2Fe-2S] cluster in recombinant Escherichia coli biotin synthase.
Jameson GN, Cosper MM, Hernández HL, Johnson MK, Huynh BH. Jameson GN, et al. Biochemistry. 2004 Feb 24;43(7):2022-31. doi: 10.1021/bi035666v. Biochemistry. 2004. PMID: 14967042 - The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase.
Jarrett JT. Jarrett JT. Arch Biochem Biophys. 2005 Jan 1;433(1):312-21. doi: 10.1016/j.abb.2004.10.003. Arch Biochem Biophys. 2005. PMID: 15581586 Review. - Biotin synthase: insights into radical-mediated carbon-sulfur bond formation.
Fugate CJ, Jarrett JT. Fugate CJ, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1213-22. doi: 10.1016/j.bbapap.2012.01.010. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22326745 Review.
Cited by
- SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly.
Ollagnier-de-Choudens S, Sanakis Y, Fontecave M. Ollagnier-de-Choudens S, et al. J Biol Inorg Chem. 2004 Oct;9(7):828-38. doi: 10.1007/s00775-004-0581-9. Epub 2004 Jul 24. J Biol Inorg Chem. 2004. PMID: 15278785 - Sulfur Administration in Fe-S Cluster Homeostasis.
Rydz L, Wróbel M, Jurkowska H. Rydz L, et al. Antioxidants (Basel). 2021 Oct 29;10(11):1738. doi: 10.3390/antiox10111738. Antioxidants (Basel). 2021. PMID: 34829609 Free PMC article. Review. - Anaerobic functionalization of unactivated C-H bonds.
Booker SJ. Booker SJ. Curr Opin Chem Biol. 2009 Feb;13(1):58-73. doi: 10.1016/j.cbpa.2009.02.036. Epub 2009 Mar 16. Curr Opin Chem Biol. 2009. PMID: 19297239 Free PMC article. Review. - A complex between biotin synthase and the iron-sulfur cluster assembly chaperone HscA that enhances in vivo cluster assembly.
Reyda MR, Fugate CJ, Jarrett JT. Reyda MR, et al. Biochemistry. 2009 Nov 17;48(45):10782-92. doi: 10.1021/bi901393t. Biochemistry. 2009. PMID: 19821612 Free PMC article. - Overexpression of biotin synthase and biotin ligase is required for efficient generation of sulfur-35 labeled biotin in E. coli.
Delli-Bovi TA, Spalding MD, Prigge ST. Delli-Bovi TA, et al. BMC Biotechnol. 2010 Oct 11;10:73. doi: 10.1186/1472-6750-10-73. BMC Biotechnol. 2010. PMID: 20937134 Free PMC article.
References
- Cohen G, Flint DH. Sanyal I, editor. Biochemistry. 1994;33:3625–3631. - PubMed
- Birch OM, Fuhrmann M, Shaw NM. J. Biol. Chem. 1995;270:19158–19165. - PubMed
- Ifuku O, Koga N, Haze S, Kishimoto J, Wachi Y. Eur. J. Biochem. 1994;224:173–178. - PubMed
- Sanyal I, Gibson KJ, Flint DH. Arch. Biochem. Biophys. 1996;326:48–56. - PubMed
- Florentin D, Bui BT, Marquet A, Ohshiro T, Izumi Y. (Ser. III).C. R. Acad. Sci. 1994;317:485–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous