SSAHA: a fast search method for large DNA databases - PubMed (original) (raw)
SSAHA: a fast search method for large DNA databases
Z Ning et al. Genome Res. 2001 Oct.
Abstract
We describe an algorithm, SSAHA (Sequence Search and Alignment by Hashing Algorithm), for performing fast searches on databases containing multiple gigabases of DNA. Sequences in the database are preprocessed by breaking them into consecutive k-tuples of k contiguous bases and then using a hash table to store the position of each occurrence of each k-tuple. Searching for a query sequence in the database is done by obtaining from the hash table the "hits" for each k-tuple in the query sequence and then performing a sort on the results. We discuss the effect of the tuple length k on the search speed, memory usage, and sensitivity of the algorithm and present the results of computational experiments which show that SSAHA can be three to four orders of magnitude faster than BLAST or FASTA, while requiring less memory than suffix tree methods. The SSAHA algorithm is used for high-throughput single nucleotide polymorphism (SNP) detection and very large scale sequence assembly. Also, it provides Web-based sequence search facilities for Ensembl projects.
Figures
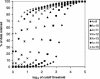
Figure 1
Percentage of data remaining as the cutoff threshold N is varied, for different values of the word size k.
Similar articles
- Refined repetitive sequence searches utilizing a fast hash function and cross species information retrievals.
Reneker J, Shyu CR. Reneker J, et al. BMC Bioinformatics. 2005 May 3;6:111. doi: 10.1186/1471-2105-6-111. BMC Bioinformatics. 2005. PMID: 15869708 Free PMC article. - muBLASTP: database-indexed protein sequence search on multicore CPUs.
Zhang J, Misra S, Wang H, Feng WC. Zhang J, et al. BMC Bioinformatics. 2016 Nov 4;17(1):443. doi: 10.1186/s12859-016-1302-4. BMC Bioinformatics. 2016. PMID: 27809763 Free PMC article. - SST: an algorithm for finding near-exact sequence matches in time proportional to the logarithm of the database size.
Giladi E, Walker MG, Wang JZ, Volkmuth W. Giladi E, et al. Bioinformatics. 2002 Jun;18(6):873-7. doi: 10.1093/bioinformatics/18.6.873. Bioinformatics. 2002. PMID: 12075023 - The EMBL Nucleotide Sequence Database. Contributing and accessing data.
Hingamp P, van den Broek AE, Stoesser G, Baker W. Hingamp P, et al. Mol Biotechnol. 1999 Oct;12(3):255-67. doi: 10.1385/MB:12:3:255. Mol Biotechnol. 1999. PMID: 10631682 Review. - Review of alignment and SNP calling algorithms for next-generation sequencing data.
Mielczarek M, Szyda J. Mielczarek M, et al. J Appl Genet. 2016 Feb;57(1):71-9. doi: 10.1007/s13353-015-0292-7. Epub 2015 Jun 9. J Appl Genet. 2016. PMID: 26055432 Review.
Cited by
- Identification of a novel polyomavirus from a marsupial host.
Dunowska M, Perrott M, Biggs P. Dunowska M, et al. Virus Evol. 2022 Oct 6;8(2):veac096. doi: 10.1093/ve/veac096. eCollection 2022. Virus Evol. 2022. PMID: 36381233 Free PMC article. - Genetic characterisation of Malawian pneumococci prior to the roll-out of the PCV13 vaccine using a high-throughput whole genome sequencing approach.
Everett DB, Cornick J, Denis B, Chewapreecha C, Croucher N, Harris S, Parkhill J, Gordon S, Carrol ED, French N, Heyderman RS, Bentley SD. Everett DB, et al. PLoS One. 2012;7(9):e44250. doi: 10.1371/journal.pone.0044250. Epub 2012 Sep 10. PLoS One. 2012. PMID: 22970189 Free PMC article. - Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens.
Park J, Zhang Y, Buboltz AM, Zhang X, Schuster SC, Ahuja U, Liu M, Miller JF, Sebaihia M, Bentley SD, Parkhill J, Harvill ET. Park J, et al. BMC Genomics. 2012 Oct 10;13:545. doi: 10.1186/1471-2164-13-545. BMC Genomics. 2012. PMID: 23051057 Free PMC article. - Programmed DNA elimination of germline development genes in songbirds.
Kinsella CM, Ruiz-Ruano FJ, Dion-Côté AM, Charles AJ, Gossmann TI, Cabrero J, Kappei D, Hemmings N, Simons MJP, Camacho JPM, Forstmeier W, Suh A. Kinsella CM, et al. Nat Commun. 2019 Nov 29;10(1):5468. doi: 10.1038/s41467-019-13427-4. Nat Commun. 2019. PMID: 31784533 Free PMC article. - New prognostic markers revealed by RNA-Seq transcriptome analysis after MYC silencing in a metastatic gastric cancer cell line.
Lopes LO, Maués JH, Ferreira-Fernandes H, Yoshioka FK, Júnior SCS, Santos AR, Ribeiro HF, Rey JA, Soares PC, Burbano RR, Pinto GR. Lopes LO, et al. Oncotarget. 2019 Oct 8;10(56):5768-5779. doi: 10.18632/oncotarget.27208. eCollection 2019 Oct 8. Oncotarget. 2019. PMID: 31645899 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. - PubMed
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials