Deletion of calcineurin and myocyte enhancer factor 2 (MEF2) binding domain of Cabin1 results in enhanced cytokine gene expression in T cells - PubMed (original) (raw)
Deletion of calcineurin and myocyte enhancer factor 2 (MEF2) binding domain of Cabin1 results in enhanced cytokine gene expression in T cells
C Esau et al. J Exp Med. 2001.
Abstract
Cabin1 binds calcineurin and myocyte enhancer factor 2 (MEF2) through its COOH-terminal region. In cell lines, these interactions were shown to inhibit calcineurin activity after T cell receptor (TCR) signaling and transcriptional activation of Nur77 by MEF2. The role of these interactions under physiological conditions was investigated using a mutant mouse strain that expresses a truncated Cabin1 lacking the COOH-terminal calcineurin and MEF2 binding domains. T and B cell development and thymocyte apoptosis were normal in mutant mice. In response to anti-CD3 stimulation, however, mutant T cells expressed significantly higher levels of interleukin (IL)-2, IL-4, IL-9, IL-13, and interferon gamma than wild-type T cells. The enhanced cytokine gene expression was not associated with change in nuclear factor of activated T cells (NF-AT)c or NF-ATp nuclear translocation but was preceded by the induction of a phosphorylated form of MEF2D in mutant T cells. Consistent with the enhanced cytokine expression, mutant mice had elevated levels of serum immunoglobulin (Ig)G1, IgG2b, and IgE and produced more IgG1 in response to a T cell-dependent antigen. These findings suggest that the calcineurin and MEF2 binding domain of Cabin1 is dispensable for thymocyte development and apoptosis, but is required for proper regulation of T cell cytokine expression probably through modulation of MEF2 activity.
Figures
Figure 1.
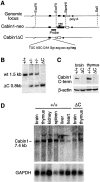
Generation of Cabin1ΔC mice. (A) Schematic diagrams of the 3′ region of the murine Cabin1 genomic locus and the Cabin1-neo and Cabin1ΔC alleles. Filled box, coding sequence; open box, 3′ untranslated region; triangle, loxP site; and filled circle, poly A site. DNA sequences at the exon-intron boundary on the Cabin1ΔC allele are shown. Capital letters refer to nucleotides from the exon and lowercase refers to nucleotides from the intron. Reading frames and the in frame stop codon (bold) 11 nucleotides into the intronic sequence are indicated. (B) Southern blot analysis for Cabin1ΔC deletion. Tail DNA was digested with BamHI and the filter was hybridized with a 0.8 kb BamHI-EcoRI probe as diagrammed in A. DNA fragments generated from the wild-type (wt) and Cabin1ΔC (ΔC) allele are indicated. (C) Western blot analysis for Cabin1 protein. Equal amount of lysates from brain and thymus of Cabin1ΔC and wild-type mice were immunoprecipitated with polyclonal antibodies specific for the COOH-terminal 77 amino acid residues of Cabin1. Precipitates were separated on SDS-PAGE, transferred to nitrocellulose membrane, and probed with the anti-Cabin1 polyclonal antibodies. Anti-actin monoclonal antibody was used for the detection of actin as a control. (D) Northern blot analysis for Cabin1 transcripts. 5 μg of poly(A)+ RNA from various tissues of wild-type and Cabin1ΔC mice were fractionated by formaldehyde-agarose gel and the filters was hybridized with a 1.2 kb Cabin1 cDNA fragment located 5′ to the deleted Cabin1 sequence. The same filter was stripped and rehybridized with a GAPDH probe.
Figure 2.
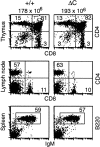
T and B cell development proceeds normally in Cabin1ΔC mice. Flow cytometric analysis of lymphoid tissues from Cabin1ΔC mice (ΔC) and wild-type controls (+/+). The numbers above the plots are the average number of thymocytes in Cabin1ΔC mice (n = 5) and wild-type littermates (n = 4). Thymocytes, lymph node cells, or splenocytes were stained with the indicated antibodies, and the numbers indicate the percentages of cells within the quadrants or gated area.
Figure 3.
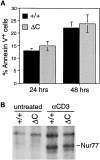
TCR-mediated apoptosis and induction of Nur77 in thymocytes of Cabin1ΔC and wild-type mice. (A) Thymocytes from 6-wk-old Cabin1ΔC mice (n = 5) and wild-type littermates (n = 4) were stimulated individually in vitro with 50 μg/ml plate-bound anti-CD3 for the indicated times. Annexin V binding of thymocytes was measured by flow cytometry. The percentages shown are after subtraction of background (Annexin V+ thymocytes in unstimulated cultures). (B) Pooled thymocytes from 6-wk-old Cabin1ΔC mice and wild-type littermates were stimulated as above for 3 h. Cell lysates containing equal amounts of protein were immunoprecipitated with anti-Nur77. Precipitates were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with the same anti-Nur77 antibody.
Figure 4.
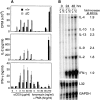
Enhanced cytokine production by Cabin1ΔC T cells. (A) Purified CD4 T cells from Cabin1ΔC (ΔC) and wild-type (+/+) mice were either not treated, or treated with indicated concentrations of anti-CD3, or 5 ng/ml of PMA plus the indicated concentrations of ionomycin for 48 h. T cell proliferation (top panel) was measured by pulsing the cultures with [3H]thymidine for 18 h after initial stimulation. The levels of IL-2 (middle panel) and IFN-γ (bottom panel) were measured in the culture supernatants after 48 h of stimulation. The assays were performed in triplicate. The proliferation and cytokine assays were also performed with total lymph node cells five and three times, respectively. Similar results were obtained except that in some experiments no difference in proliferation was observed between mutant and wild-type T cells (not shown). (B) RNase protection assay for cytokine transcripts. Lymph node cells from Cabin1ΔC and wild-type controls were stimulated for 24 and 48 h with 10 μg/ml anti-CD3, and total RNA was isolated. The levels of cytokine transcripts were assayed by RNase protection. Transcript levels were quantified by phosphorimager and normalized to the levels of transcript for the ribosomal protein L32 in each lane. The numbers indicate the fold-increase of specific cytokine transcript in Cabin1ΔC T cells over that in the wild-type T cells at 48 h. The entire assay was repeated three times with similar results.
Figure 5.
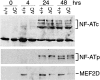
Nuclear translocation of NF-ATc and NF-ATp and induction of MEF2D. Purified T cells from Cabin1ΔC mice (ΔC) and wild-type controls (+/+) were either not stimulated or stimulated with plate-bound anti-CD3 for 4, 24, and 48 h. Nuclear extracts containing equal amounts of protein (50 μg) were separated by SDS-PAGE, transferred to membrane, and probed sequentially with antibodies specific for NF-ATc, NF-ATp, and MEF2D.
Figure 6.
Serum Ig levels are elevated in Cabin1ΔC mice. The levels of Ig isotypes in the serum of 3-mo-old Cabin1ΔC and wild-type mice were measured by ELISA. Each symbol represents one mouse. An unpaired Student's t test was used to determine the P values.
Figure 7.
Enhanced IgG1 antibody response to NP-KLH in Cabin1ΔC mice. Mice at 8–12 wk of age were immunized intravenously with NP-KLH in PBS on day 0 and 21 and bled on day 14 and 28. The levels of NP-specific Ig isotypes were measured by ELISA with NP-BSA coated plates. OD450nm at the linear range of serum dilutions is shown and each symbol represents one mouse. For the primary response, serum dilution of 1/750 was used for all isotypes and for secondary response, serum dilutions for IgG1, 2a, 2b, and 3 were 1/125,000, 1/25,000, 1/5,000, and 1/25,000, respectively. An unpaired Student's t test was used to determine the P values.
Figure 8.
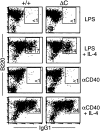
Class switching to IgG1 is unaffected in Cabin1ΔC B cells. Splenocytes from wild-type and Cabin1ΔC mice were stimulated in vitro for 5 d with LPS or anti-CD40 in the presence or absence of IL-4. Cells were stained with antibodies specific for IgG1 and B220, and analyzed by flow cytometry. Splenocytes from three wild-type and three Cabin1ΔC mice were assayed, and one representative plot for each stimulation is shown. The numbers indicate percentages of B220+ cells that are IgG1+.
Similar articles
- Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2.
Youn HD, Sun L, Prywes R, Liu JO. Youn HD, et al. Science. 1999 Oct 22;286(5440):790-3. doi: 10.1126/science.286.5440.790. Science. 1999. PMID: 10531067 - The role of NF-AT transcription factors in T cell activation and differentiation.
Serfling E, Berberich-Siebelt F, Chuvpilo S, Jankevics E, Klein-Hessling S, Twardzik T, Avots A. Serfling E, et al. Biochim Biophys Acta. 2000 Oct 20;1498(1):1-18. doi: 10.1016/s0167-4889(00)00082-3. Biochim Biophys Acta. 2000. PMID: 11042346 Review. - [The role of calcineurin in the regulation of transcription factors].
Omori N, Shibasaki F. Omori N, et al. Tanpakushitsu Kakusan Koso. 1998 Jun;43(8 Suppl):1047-54. Tanpakushitsu Kakusan Koso. 1998. PMID: 9655962 Review. Japanese. No abstract available.
Cited by
- Gene expression analyses reveal differences in children's response to malaria according to their age.
Tebben K, Yirampo S, Coulibaly D, Koné AK, Laurens MB, Stucke EM, Dembélé A, Tolo Y, Traoré K, Niangaly A, Berry AA, Kouriba B, Plowe CV, Doumbo OK, Lyke KE, Takala-Harrison S, Thera MA, Travassos MA, Serre D. Tebben K, et al. bioRxiv [Preprint]. 2023 Oct 26:2023.10.24.563751. doi: 10.1101/2023.10.24.563751. bioRxiv. 2023. PMID: 37961701 Free PMC article. Updated. Preprint. - Transcriptional defects and reprogramming barriers in somatic cell nuclear reprogramming as revealed by single-embryo RNA sequencing.
Liu Y, Wu F, Zhang L, Wu X, Li D, Xin J, Xie J, Kong F, Wang W, Wu Q, Zhang D, Wang R, Gao S, Li W. Liu Y, et al. BMC Genomics. 2018 Oct 10;19(1):734. doi: 10.1186/s12864-018-5091-1. BMC Genomics. 2018. PMID: 30305014 Free PMC article. - MEF-2 isoforms' (A-D) roles in development and tumorigenesis.
Madugula K, Mulherkar R, Khan ZK, Chigbu DI, Patel D, Harhaj EW, Jain P. Madugula K, et al. Oncotarget. 2019 Apr 12;10(28):2755-2787. doi: 10.18632/oncotarget.26763. eCollection 2019 Apr 12. Oncotarget. 2019. PMID: 31105874 Free PMC article. Review. - MEF2D sustains activation of effector Foxp3+ Tregs during transplant survival and anticancer immunity.
Di Giorgio E, Wang L, Xiong Y, Akimova T, Christensen LM, Han R, Samanta A, Trevisanut M, Bhatti TR, Beier UH, Hancock WW. Di Giorgio E, et al. J Clin Invest. 2020 Dec 1;130(12):6242-6260. doi: 10.1172/JCI135486. J Clin Invest. 2020. PMID: 32790649 Free PMC article. - Myocyte enhancer factor (MEF)-2 plays essential roles in T-cell transformation associated with HTLV-1 infection by stabilizing complex between Tax and CREB.
Jain P, Lavorgna A, Sehgal M, Gao L, Ginwala R, Sagar D, Harhaj EW, Khan ZK. Jain P, et al. Retrovirology. 2015 Feb 27;12:23. doi: 10.1186/s12977-015-0140-1. Retrovirology. 2015. PMID: 25809782 Free PMC article.
References
- Klee, C.B., H. Ren, and X. Wang. 1998. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273:13367–13370. - PubMed
- Rusnak, F., and P. Mertz. 2000. Calcineurin: form and function. Physiol. Rev. 80:1483–1521. - PubMed
- Crabtree, G.R. 2001. Calcium, calcineurin and the control of transcription. J. Biol. Chem. 276:2313–2316. - PubMed
- Friedman, J., and I. Weissman. 1991. Two cytoplasmic candidates for immunophilin action are revealed by affinity for a new cyclophilin: one in the presence and one in the absence of CsA. Cell. 66:799–806. - PubMed
- Liu, J., J.D. Farmer, Jr., W.S. Lane, J. Friedman, I. Weissman, and S.L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 66:807–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous