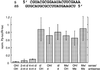Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate - PubMed (original) (raw)
Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate
S M Elbashir et al. EMBO J. 2001.
Abstract
Duplexes of 21-23 nucleotide (nt) RNAs are the sequence-specific mediators of RNA interference (RNAi) and post-transcriptional gene silencing (PTGS). Synthetic, short interfering RNAs (siRNAs) were examined in Drosophila melanogaster embryo lysate for their requirements regarding length, structure, chemical composition and sequence in order to mediate efficient RNAi. Duplexes of 21 nt siRNAs with 2 nt 3' overhangs were the most efficient triggers of sequence-specific mRNA degradation. Substitution of one or both siRNA strands by 2'-deoxy or 2'-O-methyl oligonucleotides abolished RNAi, although multiple 2'-deoxynucleotide substitutions at the 3' end of siRNAs were tolerated. The target recognition process is highly sequence specific, but not all positions of a siRNA contribute equally to target recognition; mismatches in the centre of the siRNA duplex prevent target RNA cleavage. The position of the cleavage site in the target RNA is defined by the 5' end of the guide siRNA rather than its 3' end. These results provide a rational basis for the design of siRNAs in future gene targeting experiments.
Figures
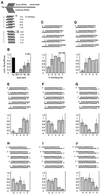
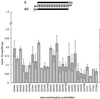
Fig. 1. Variation of the 3′ overhang of duplexes of 21 nt siRNAs. (A) Outline of the experimental strategy. The capped and polyadenylated sense target mRNA is depicted and the relative positions of sense and antisense siRNAs are shown. Eight series of duplexes according to the eight different antisense strands were prepared. The siRNA sequences and the number of overhanging nucleotides were changed in 1 nt steps. (B) Normalized relative luminescence of target luciferase (Photinus pyralis, Pp-luc) to control luciferase (Renilla reniformis, Rr-luc) in D.melanogaster embryo lysate in the presence of 5 nM blunt-ended dsRNAs (Elbashir et al., 2001b). The luminescence ratios determined in the presence of dsRNA were normalized to the ratio obtained for a buffer control (bu; black bar). Normalized ratios less than 1 indicate specific interference. (C–J) Normalized interference ratios for eight series of 21 nt siRNA duplexes. The sequences of siRNA duplexes are depicted above the bar graphs. Each part shows the interference ratio for a set of duplexes formed with a given antisense guide siRNA and five different sense siRNAs. The number of overhanging nucleotides (3′ overhang, positive numbers; 5′ overhang, negative numbers) is indicated on the _x_-axis. Data points were averaged from at least three independent experiments. Error bars represent standard deviations.
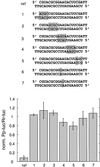
Fig. 2. Variation of the length of the sense strand of siRNA duplexes. (A) Representation of the experiment. Three 21 nt antisense strands were paired with eight sense siRNAs. The siRNAs were changed in length at their 3′ end. The 3′ overhang of the antisense siRNA was 1 nt (B), 2 nt (C) or 3 nt (D), while the sense siRNA overhang was varied for each series. The sequences of the siRNA duplexes and the corresponding interference ratios are indicated.
Fig. 3. Variation of the length of siRNA duplexes with preserved 2 nt 3′ overhangs. (A) Graphic representation of the experiment. The 21 nt siRNA duplex is identical in sequence to the one shown in Figures 1H and 2C. The siRNA duplexes were extended to the 3′ side of the sense siRNA (B) or the 5′ side of the sense siRNA (C). The siRNA duplex sequences and the respective interference ratios are indicated.
Fig. 4. Substitution of the 2′-hydroxyl groups of the siRNA ribose residues. The 2′-hydroxyl groups (OH) in the strands of siRNA duplexes were replaced by 2′-deoxy (d) or 2′-_O_-methyl (Me). 2 and 4 nt 2′-deoxy substitutions at the 3′ ends are indicated as 2- and 4-nt d, respectively. Uridine residues were replaced by 2′-deoxythymidine.
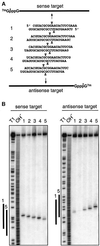
Fig. 5. Mapping of sense and antisense target RNA cleavage by 21 nt siRNA duplexes with 2 nt 3′ overhangs. (A) Representation of 32P (asterisk) cap-labelled sense and antisense target RNAs and siRNA duplexes. The position of sense and antisense target RNA cleavage is indicated by triangles on top and below the siRNA duplexes, respectively. (B) Mapping of target RNA cleavage sites. After 2 h incubation of 10 nM target RNA with 100 nM siRNA duplex in D.melanogaster embryo lysate, the 5′ cap-labelled substrate and the 5′ cleavage products were resolved on 6% sequencing gels. Length markers were generated by partial RNase T1 digestion (T1) and partial alkaline hydrolysis (OH) of the target RNAs. The bold lines to the left of the images indicate the region covered by the siRNA strands 1 and 5 of the same orientation as the target.
Fig. 6. The 5′ end of a guide siRNA defines the position of target RNA cleavage. (A and B) Representation of the experimental strategy. The antisense siRNA was the same in all siRNA duplexes, but the sense strand was varied between 18 and 25 nt by changing the 3′ end (A) or 18 and 23 nt by changing the 5′ end (B). The position of sense and antisense target RNA cleavage is indicated by triangles on top and below the siRNA duplexes, respectively. (C and D) Analysis of target RNA cleavage using cap-labelled sense (top) or antisense (bottom) target RNAs. The residual amount of targeted substrate and the cap-labelled 5′ cleavage products are shown. The sequences of the siRNA duplexes are indicated and the length of the sense siRNA strands is marked on top. The control lane, marked with a dash in (C), shows target RNA incubated in absence of siRNAs. Markers were as described in Figure 5. The arrows in (D), bottom, indicate the target RNA cleavage sites that differ by 1 nt.
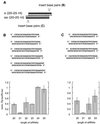
Fig. 7. Sequence variation of the 3′ overhang of siRNA duplexes. The 2 nt 3′ overhang (NN, in grey) was changed in sequence and composition as indicated (T, 2′-deoxythymidine; dG, 2′-deoxyguanosine; asterisk, wild-type siRNA duplex). Normalized interference ratios were determined as described in Figure 1. The wild-type sequence is the same as depicted in Figure 4.
Fig. 8. Sequence specificity of target recognition. The sequences of the mismatched siRNA duplexes are shown, modified sequence segments or single nucleotides are shaded in grey. The reference duplex (ref) and the siRNA duplexes 1–7 contain 2′-deoxythymidine 2 nt overhangs. The silencing efficiency of the thymidine-modified reference duplex was comparable to the wild-type sequence (Figure 7). Normalized interference ratios were determined as described in Figure 1.
Similar articles
- RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs.
Lipardi C, Wei Q, Paterson BM. Lipardi C, et al. Cell. 2001 Nov 2;107(3):297-307. doi: 10.1016/s0092-8674(01)00537-2. Cell. 2001. PMID: 11701121 - RNA interference is mediated by 21- and 22-nucleotide RNAs.
Elbashir SM, Lendeckel W, Tuschl T. Elbashir SM, et al. Genes Dev. 2001 Jan 15;15(2):188-200. doi: 10.1101/gad.862301. Genes Dev. 2001. PMID: 11157775 Free PMC article. - Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing.
Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Harborth J, et al. Antisense Nucleic Acid Drug Dev. 2003 Apr;13(2):83-105. doi: 10.1089/108729003321629638. Antisense Nucleic Acid Drug Dev. 2003. PMID: 12804036 - Gene silencing mediated by small interfering RNAs in mammalian cells.
Scherr M, Morgan MA, Eder M. Scherr M, et al. Curr Med Chem. 2003 Feb;10(3):245-56. doi: 10.2174/0929867033368493. Curr Med Chem. 2003. PMID: 12570711 Review. - A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst.
Nishikura K. Nishikura K. Cell. 2001 Nov 16;107(4):415-8. doi: 10.1016/s0092-8674(01)00581-5. Cell. 2001. PMID: 11719182 Review.
Cited by
- Structure of yeast Argonaute with guide RNA.
Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Nakanishi K, et al. Nature. 2012 Jun 20;486(7403):368-74. doi: 10.1038/nature11211. Nature. 2012. PMID: 22722195 Free PMC article. - MicroRNAs: molecular features and role in cancer.
Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. Lages E, et al. Front Biosci (Landmark Ed). 2012 Jun 1;17(7):2508-40. doi: 10.2741/4068. Front Biosci (Landmark Ed). 2012. PMID: 22652795 Free PMC article. Review. - Stability of miRNA 5'terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy.
Hibio N, Hino K, Shimizu E, Nagata Y, Ui-Tei K. Hibio N, et al. Sci Rep. 2012;2:996. doi: 10.1038/srep00996. Epub 2012 Dec 18. Sci Rep. 2012. PMID: 23251782 Free PMC article. - Evaluation of canonical siRNA and Dicer substrate RNA for inhibition of hepatitis C virus genome replication--a comparative study.
Carneiro B, Braga AC, Batista MN, Harris M, Rahal P. Carneiro B, et al. PLoS One. 2015 Feb 23;10(2):e0117742. doi: 10.1371/journal.pone.0117742. eCollection 2015. PLoS One. 2015. PMID: 25705875 Free PMC article. - In vitro characterization of the activity of PF-05095808, a novel biological agent for hepatitis C virus therapy.
Lavender H, Brady K, Burden F, Delpuech-Adams O, Denise H, Palmer A, Perkins H, Savic B, Scott S, Smith-Burchnell C, Troke P, Wright JF, Suhy D, Corbau R. Lavender H, et al. Antimicrob Agents Chemother. 2012 Mar;56(3):1364-75. doi: 10.1128/AAC.05357-11. Epub 2011 Dec 27. Antimicrob Agents Chemother. 2012. PMID: 22203606 Free PMC article.
References
- Ambros V. (2000) Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev., 10, 428–433. - PubMed
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. - PubMed
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. - PubMed
- Carthew R.W. (2001) Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol., 13, 244–248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials