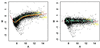Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation - PubMed (original) (raw)
Comparative Study
Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation
Yee Hwa Yang et al. Nucleic Acids Res. 2002.
Abstract
There are many sources of systematic variation in cDNA microarray experiments which affect the measured gene expression levels (e.g. differences in labeling efficiency between the two fluorescent dyes). The term normalization refers to the process of removing such variation. A constant adjustment is often used to force the distribution of the intensity log ratios to have a median of zero for each slide. However, such global normalization approaches are not adequate in situations where dye biases can depend on spot overall intensity and/or spatial location within the array. This article proposes normalization methods that are based on robust local regression and account for intensity and spatial dependence in dye biases for different types of cDNA microarray experiments. The selection of appropriate controls for normalization is discussed and a novel set of controls (microarray sample pool, MSP) is introduced to aid in intensity-dependent normalization. Lastly, to allow for comparisons of expression levels across slides, a robust method based on maximum likelihood estimation is proposed to adjust for scale differences among slides.
Figures
Figure 1
Within-slide normalization. (A) MA-plot demonstrating the need for within-print tip group location normalization. (B) MA-plot after within-print tip group location normalization. Both panels display the lowess fits (f = 40%) for each of the 16 print tip groups (data from apo AI knockout mouse number 8 in experiment A).
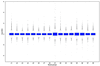
Figure 2
Within-slide normalization: box plots displaying the intensity log ratio distribution, for each of the 16 print tip groups before and after different normalization procedures. The array was printed using a 4 × 4 print head and the print tip groups are numbered first from left to right, then from top to bottom, starting from the top left corner (data from apo AI knockout mouse number 8 in experiment A). (A) Before normalization. (B) After within-print tip group location normalization, but before scale adjustment. (C) After within-print tip group location and scale normalization.
Figure 3
Within-slide normalization: MA-plot for comparison of the anterior versus posterior portion of the olfactory bulb. These samples are very similar and we do not expect many genes to change. The cyan dots represent the MSP titration series and the cyan curve represents the corresponding lowess fit. The red curve corresponds to the lowess fit for the entire dataset. Control genes are highlighted in yellow (tubulin and GAPDH), green (mouse genomic DNA) and orange (an approximate rank-invariant set of genes with P = 0.01 and l = 25). (Left) MA-plot before normalization. (Right) MA-plot after within-print tip group location normalization.
Figure 4
Within-slide normalization: MA-plot for comparison of the medial versus lateral portion of the olfactory bulb. The cyan dots represent the MSP titration series and the cyan curve represents the corresponding lowess fit. The red curve corresponds to the lowess fit for the entire dataset. The green curve represents the composite normalization curve. Control genes are highlighted in yellow (tubulin and GAPDH), green (mouse genomic DNA) and orange (an approximate rank-invariant set of genes). (A) MA-plot before normalization. (B) MA-plot after composite normalization.
Figure 5
Within-slide normalization. (A) Density plots of the log ratios M before and after different normalization procedures. The solid black curve represents the density of the log ratios before normalization. The red, green, blue and cyan curves represent the densities after global median normalization, intensity-dependent location normalization, within-print tip group location normalization and within-print tip group scale normalization, respectively (data from apo AI knockout mouse number 8 in experiment A). (B) Plot of _t_-statistics for different normalization methods. The numbers 1–8 represent the differentially expressed genes identified in Dudoit et al. (10) and confirmed using RT–PCR: indices 1–3 represent the three apo AI genes spotted on the array. Empty circles represent the remaining 6376 genes where no effect is expected. Only t values less than –4 are shown.
Figure 6
Multiple slide normalization: box plots displaying the intensity log ratio distribution for different slides/mice for experiment A, after within-print tip group location and scale normalization. The first eight box plots represent the data for the eight control mice and the last eight represent the data for the eight apo AI knockout mice.
Similar articles
- A robust neural networks approach for spatial and intensity-dependent normalization of cDNA microarray data.
Tarca AL, Cooke JE, Mackay J. Tarca AL, et al. Bioinformatics. 2005 Jun 1;21(11):2674-83. doi: 10.1093/bioinformatics/bti397. Epub 2005 Mar 29. Bioinformatics. 2005. PMID: 15797913 - A new non-linear normalization method for reducing variability in DNA microarray experiments.
Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. Workman C, et al. Genome Biol. 2002 Aug 30;3(9):research0048. doi: 10.1186/gb-2002-3-9-research0048. Epub 2002 Aug 30. Genome Biol. 2002. PMID: 12225587 Free PMC article. - Normalization for two-channel microarray data.
Ittrich C. Ittrich C. Methods Inf Med. 2005;44(3):418-22. Methods Inf Med. 2005. PMID: 16113767 - Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available.
Benes V, Castoldi M. Benes V, et al. Methods. 2010 Apr;50(4):244-9. doi: 10.1016/j.ymeth.2010.01.026. Epub 2010 Jan 28. Methods. 2010. PMID: 20109550 Review. - Statistical tests for differential expression in cDNA microarray experiments.
Cui X, Churchill GA. Cui X, et al. Genome Biol. 2003;4(4):210. doi: 10.1186/gb-2003-4-4-210. Epub 2003 Mar 17. Genome Biol. 2003. PMID: 12702200 Free PMC article. Review.
Cited by
- Butyrate induces higher host transcriptional changes to inhibit porcine epidemic diarrhea virus strain CV777 infection in porcine intestine epithelial cells.
Zhong Z, Zhang Y, Zhao X, Zhou C, Zhu S, Wu J. Zhong Z, et al. Virol J. 2024 Jul 11;21(1):157. doi: 10.1186/s12985-024-02428-5. Virol J. 2024. PMID: 38992629 Free PMC article. - A systems genomics and genetics approach to identify the genetic regulatory network for lignin content in Brassica napus seeds.
Zhang W, Higgins EE, Robinson SJ, Clarke WE, Boyle K, Sharpe AG, Fobert PR, Parkin IAP. Zhang W, et al. Front Plant Sci. 2024 Jun 5;15:1393621. doi: 10.3389/fpls.2024.1393621. eCollection 2024. Front Plant Sci. 2024. PMID: 38903439 Free PMC article. - A framework for performance enhancement of classifiers in detection of prostate cancer from microarray gene.
Mani K, Rajaguru H. Mani K, et al. Heliyon. 2024 Apr 25;10(9):e29630. doi: 10.1016/j.heliyon.2024.e29630. eCollection 2024 May 15. Heliyon. 2024. PMID: 38720727 Free PMC article. - Removing unwanted variation between samples in Hi-C experiments.
Fletez-Brant K, Qiu Y, Gorkin DU, Hu M, Hansen KD. Fletez-Brant K, et al. Brief Bioinform. 2024 Mar 27;25(3):bbae217. doi: 10.1093/bib/bbae217. Brief Bioinform. 2024. PMID: 38711367 Free PMC article. - Label-Free Quantitation of Endogenous Peptides.
Abid MSR, Qiu H, Checco JW. Abid MSR, et al. Methods Mol Biol. 2024;2758:125-150. doi: 10.1007/978-1-0716-3646-6_7. Methods Mol Biol. 2024. PMID: 38549012 Free PMC article.
References
- Taniguchi M., Miura,K., Iwao,H. and Yamanaka,S. (2001) Quantitative assessment of DNA microarrays—comparison with northern blot analyses. Genomics, 71, 34–39. - PubMed
- Hughes T.R., Marton,M.J., Jones,A.R., Roberts,C.J., Stoughton,R., Armour,C.D., Bennett,H.A., Coffey,E., Dai,H., He,Y.D., Kidd,M.J., King,A.M., Meyer,M.R., Slade,D., Lum,P.Y., Stepaniants,S.B., Shoemaker,D.D., Gachotte,D., Chakraburtty,K., Simon,J., Bard,M. and Friend,S.H. (2000) Functional discovery via a compendium of expression profiles. Cell, 102, 109–126. - PubMed
- Alizadeh A.A., Eisen,M.B., Davis,R.E., Ma,C., Lossos,I.S., Rosenwald,A., Boldrick,J.C., Sabet,H., Tran,T., Yu,X., Powell,J.I., Yang,L., Marti,G.E., Moore,T., Hudson,J.,Jr, Lu,L., Lewis,D.B., Tibshirani,R., Sherlock,G., Chan,W.C., Greiner,T.C., Weisenburger,D.D., Armitage,J.O., Warnke,R., Staudt,L.M. et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature, 403, 503–511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials