Discovery of five conserved beta -defensin gene clusters using a computational search strategy - PubMed (original) (raw)
Discovery of five conserved beta -defensin gene clusters using a computational search strategy
Brian C Schutte et al. Proc Natl Acad Sci U S A. 2002.
Erratum in
- Proc Natl Acad Sci U S A 2002 Oct 29;99(22):14611
Abstract
The innate immune system includes antimicrobial peptides that protect multicellular organisms from a diverse spectrum of microorganisms. beta-Defensins comprise one important family of mammalian antimicrobial peptides. The annotation of the human genome fails to reveal the expected diversity, and a recent query of the draft sequence with the blast search engine found only one new beta-defensin gene (DEFB3). To define better the beta-defensin gene family, we adopted a genomics approach that uses hmmer, a computational search tool based on hidden Markov models, in combination with blast. This strategy identified 28 new human and 43 new mouse beta-defensin genes in five syntenic chromosomal regions. Within each syntenic cluster, the gene sequences and organization were similar, suggesting each cluster pair arose from a common ancestor and was retained because of conserved functions. Preliminary analysis indicates that at least 26 of the predicted genes are transcribed. These results demonstrate the value of a genomewide search strategy to identify genes with conserved structural motifs. Discovery of these genes represents a new starting point for exploring the role of beta-defensins in innate immunity.
Figures
Figure 1
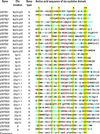
Multiple sequence alignment of human β-defensin proteins. The amino acid sequences were predicted from genomic sequence (Table 1). The chromosomal location of each gene is indicated, except where mapping was ambiguous (A). The sequences were aligned as described in Materials and Methods followed by minor manual manipulations to maximize sequence alignment and clustering of genes by chromosome. We classified β-defensin genes as known (K), if evidence exists that they are transcribed and that their protein product demonstrates antimicrobial activity; related (R), if evidence exists that they are transcribed but their protein product has not been tested for antimicrobial activity; predicted (P), if no evidence exists that they are transcribed; and pseudogenes (S), if the DNA sequence is highly similar to a β-defensin gene, but the predicted amino acid sequence lacks an ORF across the six-cysteine motif. The consensus sequence shows cysteine residues (yellow highlight), positively charged residues (blue +) and other residues (red) if they are represented in greater than 50% of all predicted β-defensin proteins.
Figure 2
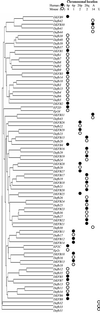
Dendrogram of predicted β-defensin proteins. The length of each branch is inversely related to their similarity. The tree was constructed with the partial amino acid sequences predicted from the indicated human (●) and mouse (○) known, related, and predicted genes (Table 1). The genomic location for each gene is indicated. In some cases, the location of the genes was ambiguous (A) or unknown (U).
Figure 3
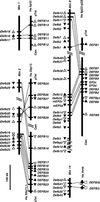
Genomic structure of five conserved β-defensin gene clusters. Vertical bars represent assembled genomic DNA sequence contigs (see Table 1 for GenBank and Celera accession nos.) from the indicated human (Hs) and mouse (Mm) chromosome. Slanted lines represent gaps in the genomic sequence of known (single) or unknown (double) length. The telomere (Tel) and centromere (Cen) orientation of human DNA sequence contigs was deduced from the position of genetic markers. The orientation of the mouse DNA sequence contigs was deduced from the alignment of human and mouse gene homologs. The direction of transcription is indicated for known and related genes (▴) and predicted genes and pseudogenes (▵) or is unknown (▫). Thin lines connect human and mouse genes with highest sequence similarity (Fig. 2).
Similar articles
- Genomics-based approaches to gene discovery in innate immunity.
Scheetz T, Bartlett JA, Walters JD, Schutte BC, Casavant TL, McCray PB Jr. Scheetz T, et al. Immunol Rev. 2002 Dec;190:137-45. doi: 10.1034/j.1600-065x.2002.19010.x. Immunol Rev. 2002. PMID: 12493011 Review. - Cross-species analysis of the mammalian beta-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract.
Patil AA, Cai Y, Sang Y, Blecha F, Zhang G. Patil AA, et al. Physiol Genomics. 2005 Sep 21;23(1):5-17. doi: 10.1152/physiolgenomics.00104.2005. Epub 2005 Jul 20. Physiol Genomics. 2005. PMID: 16033865 - Rapid evolution and diversification of mammalian alpha-defensins as revealed by comparative analysis of rodent and primate genes.
Patil A, Hughes AL, Zhang G. Patil A, et al. Physiol Genomics. 2004 Dec 15;20(1):1-11. doi: 10.1152/physiolgenomics.00150.2004. Epub 2004 Oct 19. Physiol Genomics. 2004. PMID: 15494476 - A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins.
Xiao Y, Hughes AL, Ando J, Matsuda Y, Cheng JF, Skinner-Noble D, Zhang G. Xiao Y, et al. BMC Genomics. 2004 Aug 13;5(1):56. doi: 10.1186/1471-2164-5-56. BMC Genomics. 2004. PMID: 15310403 Free PMC article. - Rapid sequence divergence in mammalian beta-defensins by adaptive evolution.
Maxwell AI, Morrison GM, Dorin JR. Maxwell AI, et al. Mol Immunol. 2003 Nov;40(7):413-21. doi: 10.1016/s0161-5890(03)00160-3. Mol Immunol. 2003. PMID: 14568387 Review.
Cited by
- Roles of human beta-defensins in innate immune defense at the ocular surface: arming and alarming corneal and conjunctival epithelial cells.
Garreis F, Schlorf T, Worlitzsch D, Steven P, Bräuer L, Jäger K, Paulsen FP. Garreis F, et al. Histochem Cell Biol. 2010 Jul;134(1):59-73. doi: 10.1007/s00418-010-0713-y. Epub 2010 Jun 5. Histochem Cell Biol. 2010. PMID: 20526610 - Antimicrobial peptide-like genes in Nasonia vitripennis: a genomic perspective.
Tian C, Gao B, Fang Q, Ye G, Zhu S. Tian C, et al. BMC Genomics. 2010 Mar 19;11:187. doi: 10.1186/1471-2164-11-187. BMC Genomics. 2010. PMID: 20302637 Free PMC article. - Human {beta}-defensin 2 is expressed and associated with Mycobacterium tuberculosis during infection of human alveolar epithelial cells.
Rivas-Santiago B, Schwander SK, Sarabia C, Diamond G, Klein-Patel ME, Hernandez-Pando R, Ellner JJ, Sada E. Rivas-Santiago B, et al. Infect Immun. 2005 Aug;73(8):4505-11. doi: 10.1128/IAI.73.8.4505-4511.2005. Infect Immun. 2005. PMID: 16040961 Free PMC article. - Expressed sequence tag profiling identifies developmental and anatomic partitioning of gene expression in the mouse prostate.
Abbott DE, Pritchard C, Clegg NJ, Ferguson C, Dumpit R, Sikes RA, Nelson PS. Abbott DE, et al. Genome Biol. 2003;4(12):R79. doi: 10.1186/gb-2003-4-12-r79. Epub 2003 Nov 28. Genome Biol. 2003. PMID: 14659016 Free PMC article. - Induction of beta defensin 2 by NTHi requires TLR2 mediated MyD88 and IRAK-TRAF6-p38MAPK signaling pathway in human middle ear epithelial cells.
Lee HY, Takeshita T, Shimada J, Akopyan A, Woo JI, Pan H, Moon SK, Andalibi A, Park RK, Kang SH, Kang SS, Gellibolian R, Lim DJ. Lee HY, et al. BMC Infect Dis. 2008 Jun 25;8:87. doi: 10.1186/1471-2334-8-87. BMC Infect Dis. 2008. PMID: 18578886 Free PMC article.
References
- Huttner K M, Bevins C L. Pediatr Res. 1999;45:785–794. - PubMed
- Tang Y Q, Selsted M E. J Biol Chem. 1993;268:6649–6653. - PubMed
- Bensch K W, Raida M, Magert H J, Schulz-Knappe P, Forssmann W G. FEBS Lett. 1995;368:331–335. - PubMed
- Harder J, Bartels J, Christophers E, Schroder J-M. Nature (London) 1997;387:861–862. - PubMed
- Harder J, Bartels J, Christophers E, Schroder J M. J Biol Chem. 2001;276:5707–5713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30-HD27748/HD/NICHD NIH HHS/United States
- CA-85188/CA/NCI NIH HHS/United States
- HL-61234/HL/NHLBI NIH HHS/United States
- K12 HD027748/HD/NICHD NIH HHS/United States
- P50 HL061234/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials