Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities - PubMed (original) (raw)
Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities
Young-Ho Lee et al. Mol Cell Biol. 2002 Jun.
Abstract
Hormone-activated nuclear receptors (NR) bind to specific regulatory DNA elements associated with their target genes and recruit coactivator proteins to remodel chromatin structure, recruit RNA polymerase, and activate transcription. The p160 coactivators (e.g., SRC-1, GRIP1, and ACTR) bind directly to activated NR and can recruit a variety of secondary coactivators. We have established a transient-transfection assay system under which the activity of various NR is highly or completely dependent on synergistic cooperation among three classes of coactivators: a p160 coactivator, the protein methyltransferase CARM1, and any of the three protein acetyltransferases, p300, CBP, or p/CAF. The three-coactivator functional synergy was only observed when low levels of NR were expressed and was highly or completely dependent on the methyltransferase activity of CARM1 and the acetyltransferase activity of p/CAF, but not the acetyltransferase activity of p300. Other members of the protein arginine methyltransferase family, which methylate different protein substrates than CARM1, could not substitute for CARM1 to act synergistically with p300 or p/CAF. A ternary complex of GRIP1, CARM1, and p300 or CBP was demonstrated in cultured mammalian cells, supporting a physiological role for the observed synergy. The transfection assay described here is a valuable new tool for investigating the mechanism of coactivator function and demonstrates the importance of multiple coactivators, including CARM1 and its specific protein methyltransferase activity, in transcriptional activation.
Figures
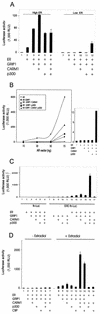
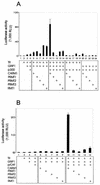
FIG. 1.
Requirement for three coactivators (GRIP1, CARM1, and p300) at low levels of NR. CV-1 cells in each well of six-well culture dishes were transiently transfected with plasmids as indicated below. The transfected cells were grown with 20 nM E2 for ER or DHT for AR. Cell extracts were prepared and assayed for luciferase activity. (A) MMTV(ERE)-LUC reporter plasmid, 250 ng; 10 ng (low ER) or 100 ng (high ER) of ER expression vector; 250 ng of pSG5.HA-GRIP1, 500 ng of pSG5.HA-CARM1, and 500 ng of pCMV-p300. (B) MMTV-LUC, 250 ng; 10 to 70 ng of AR expression vector; coactivator vectors as for panel A. The graph on the right shows the activity observed with 10 ng of AR vector on an expanded scale. (C) EREII-LUC(GL45) or TK-LUC reporter plasmid, 250 ng; 1 ng of ER expression vector; coactivator vectors as for panel A. (D) EREII-LUC(GL45), 250 ng; ER expression vector, 1 ng; 500 ng of each coactivator expression vector (GRIP1, CARM1, p300, and CBP). +, present.
FIG. 2.
Ternary coactivator complex formation among GRIP1, CARM1 and p300/CBP. (A) Coimmunoprecipitation. Cos-7 cells in 100-mm-diameter dishes were transfected with coactivator expression vectors as indicated: 2.5 μg of pCMV-p300 (produces p300 with a Flag tag); 2.5 μg of pSG5.HA-CARM1; 2.5 μg of pSG5.HA-GRIP1 (wild type [WT]) or the equivalent vector encoding GRIP1ΔAD1 (ΔAD1), GRIP1ΔAD2 (ΔAD2), or GRIP1ΔAD1 plus ΔAD2 (ΔAD1/2). Com-plexes containing p300 were immunoprecipitated from transfected cell extracts with anti-Flag antibody, and coprecipitated CARM1 was detected by immunoblotting with antibodies against the HA tag (top). To check CARM1 expression before immunoprecipitation, 5% of the transfected cell extract was directly tested by immunoblotting with antibodies against HA tag (bottom). Lanes 1 to 3 and 4 to 10 represent two independent experiments. The diagram indicates the proposed interaction sites among the three coactivators. IP, antibody used for immunoprecipitation; WB, antibody used for Western immunoblotting; IgG-H, immunoglobulin G heavy chain from the immunoprecipitation, which is recognized by the secondary antibody used in the immunoblot. (B) Modified mammalian two-hybrid system. CV-1 cells in six-well dishes were transfected with 250 ng of GK1 reporter plasmid controlled by an E1b basal promoter (E1bTATA) and Gal4 response elements (Gal4RE) and, as indicated, 250 ng of pGAL-CBP8, encoding Gal4 DBD fused to full-length CBP; 250 ng of pVP16.CARM1, encoding VP16 activation domain (VP16 AD) fused to CARM1; and 500 ng of pSG5.HA-GRIP1 or the corresponding vector expressing the ΔAD1, ΔAD2, or ΔAD1 plus ΔAD2 mutant of GRIP1. The luciferase (Luc) activity of the cell extract is shown. (C) Coactivator assays. CV1 cells in six-well dishes were transfected with 250 ng of MMTV(TRE)-LUC reporter plasmid, 1 ng of TR expression vector, 500 ng of CARM1 vector, 500 ng of p300 vector, and 250 ng of the indicated wild-type or mutant GRIP1 vector. The cells were grown with 20 nM T3, and the luciferase activity of the cell extract was determined. +, present.
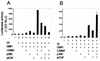
FIG. 3.
Synergy among various combinations of three or four coactivators at low NR levels. CV-1 cells were transiently transfected as for Fig. 1 and grown with 20 nM E2 for ER or 20 nM T3 for TR. The plasmids used were 250 ng of MMTV(ERE)-LUC, 1 ng of ER vector, 250 ng of GRIP1 vector, 500 ng of CARM1 vector, 500 ng of p300 vector, and 500 ng of p/CAF vector (A) and 250 ng of MMTV(TRE)-LUC, 5 ng of TR vector, and coactivator vectors as for panel A (B). +, present.
FIG. 4.
Role of protein acetyltransferase activities of p/CAF and p300 in coactivator synergy. CV-1 cells in six-well dishes were transfected with 1 ng of AR or TR expression vector, 250 ng of MMTV-LUC or MMTV(TRE)-LUC, 250 ng of GRIP1 vector, 500 ng of CARM1 vector, and 500 ng of a vector encoding the indicated wild-type or mutant form of p/CAF (A) or p300 (B). The cells were grown with 20 nM DHT or T3, and cell extracts were assayed for luciferase activity. WT, wild-type p/CAF or p300; MT1, p/CAFΔ579-608; MT2, p/CAFΔ609-624; MT, p300Δ1603-1653; +, present.
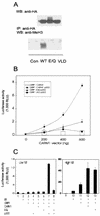
FIG. 5.
Role of protein methyltransferase activity of CARM1 in coactivator synergy. (A) Methyltransferase activities of wild-type and mutant CARM1. Cos-7 cells (100-mm-diameter dishes) were transfected with 2.5 μg of the indicated CARM1 expression vector: Con, control with pSG5.HA; WT, pSG5.HA-CARM1 wild type; E/Q, pSG5.HA-CARM1(E267Q) mutant; VLD, pSG5.HA-CARM1(VLD) mutant. Cell extracts were immunoprecipitated (IP) with antibody against HA tag (anti-HA), and immunoprecipitates were incubated with mixed histones and _S_-adenosylmethionine to allow methylation. Incubated reactions were analyzed by Western immunoblotting (WB), using antibodies specific for the CARM1-methylated form of histone H3 (anti-MeH3; bottom). Expression of CARM1 was assessed before immunoprecipitation by immunoblotting with antibodies against HA tag (top). (B) Coactivator synergy with different amounts of wild-type and mutant CARM1. CV-1 cells in six-well dishes were transfected with 1 ng of TR vector, 250 ng of MMTV(TRE)-LUC reporter plasmid, 250 ng of GRIP1 vector, 500 ng of p300 vector (line plots), or no p300 vector (bars) and the indicated amount of CARM1 wild-type ormutant vector. The cells were grown with 20 nM T3, and the luciferase activities of the cell extracts were determined. (C) Coactivator activities of mutant and wild-type CARM1 at low versus high NR levels. CV-1 cells in six-well dishes were transfected with 1 (Low NR) or 100 (High NR) ng of ER vector, 250 ng of MMTV(ERE)-LUC reporter plasmid, 250 ng of GRIP1 vector, 500 ng of p300 vector, and 500 ng of wild-type or mutant CARM1 vector. The cells were grown with 20 nM E2, and the luciferase activities of the cell extracts were determined. +, present.
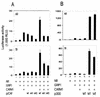
FIG. 6.
Selective synergy of protein arginine methyltransferases with p300 and p/CAF. CV-1 cells in six-well dishes were transfected with 1 ng of TR vector, 250 ng of MMTV(TRE)-LUC reporter plasmid, and coactivator vectors as indicated: 250 ng of GRIP1 vector, 500 ng of p300 (A) or p/CAF (B) vector, and 500 ng of the indicated methyltransferase vector encoding CARM1, PRMT1, PRMT2, PRMT3, or RMT1. The cells were grown with 20 nM T3, and the luciferase activities of the cell extracts were determined. +, present.
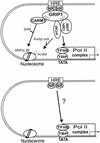
FIG. 7.
Different mechanisms of transcriptional activation at low and high NR levels. (Top) At low NR levels, NRs shuttle on and off of the hormone response element (HRE), and occupancy of the HRE is relatively infrequent. Transcriptional activation by the bound NRs requires the assistance of multiple coactivators to remodel chromatin structure and recruit and activate RNA polymerase II (Pol II complex). (Bottom) High NR levels may force almost constant occupancy of the HREs by NRs. Perhaps such high occupancy allows NRs to recruit RNA polymerase through direct contact with TATA binding protein (TBP), TFIIB, or other components of the RNA polymerase II complex, independent of the action of some or many coactivators.
Similar articles
- Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities.
Koh SS, Chen D, Lee YH, Stallcup MR. Koh SS, et al. J Biol Chem. 2001 Jan 12;276(2):1089-98. doi: 10.1074/jbc.M004228200. J Biol Chem. 2001. PMID: 11050077 - Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity.
Lee YH, Campbell HD, Stallcup MR. Lee YH, et al. Mol Cell Biol. 2004 Mar;24(5):2103-17. doi: 10.1128/MCB.24.5.2103-2117.2004. Mol Cell Biol. 2004. PMID: 14966289 Free PMC article. - Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300.
Chen D, Huang SM, Stallcup MR. Chen D, et al. J Biol Chem. 2000 Dec 29;275(52):40810-6. doi: 10.1074/jbc.M005459200. J Biol Chem. 2000. PMID: 11010967 - The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators.
Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. Stallcup MR, et al. J Steroid Biochem Mol Biol. 2003 Jun;85(2-5):139-45. doi: 10.1016/s0960-0760(03)00222-x. J Steroid Biochem Mol Biol. 2003. PMID: 12943698 Review. - The SRC family of nuclear receptor coactivators.
Leo C, Chen JD. Leo C, et al. Gene. 2000 Mar 7;245(1):1-11. doi: 10.1016/s0378-1119(00)00024-x. Gene. 2000. PMID: 10713439 Review.
Cited by
- Glucocorticoid-dependent phosphorylation of the transcriptional coregulator GRIP1.
Dobrovolna J, Chinenov Y, Kennedy MA, Liu B, Rogatsky I. Dobrovolna J, et al. Mol Cell Biol. 2012 Feb;32(4):730-9. doi: 10.1128/MCB.06473-11. Epub 2011 Dec 12. Mol Cell Biol. 2012. PMID: 22158970 Free PMC article. - Transcriptional activation of human matrix metalloproteinase-9 gene expression by multiple co-activators.
Zhao X, Benveniste EN. Zhao X, et al. J Mol Biol. 2008 Nov 28;383(5):945-56. doi: 10.1016/j.jmb.2008.08.071. Epub 2008 Sep 4. J Mol Biol. 2008. PMID: 18790699 Free PMC article. - Minireview: nuclear receptor coregulators of the p160 family: insights into inflammation and metabolism.
Rollins DA, Coppo M, Rogatsky I. Rollins DA, et al. Mol Endocrinol. 2015 Apr;29(4):502-17. doi: 10.1210/me.2015-1005. Epub 2015 Feb 3. Mol Endocrinol. 2015. PMID: 25647480 Free PMC article. Review. - Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus.
Hu SB, Xiang JF, Li X, Xu Y, Xue W, Huang M, Wong CC, Sagum CA, Bedford MT, Yang L, Cheng D, Chen LL. Hu SB, et al. Genes Dev. 2015 Mar 15;29(6):630-45. doi: 10.1101/gad.257048.114. Genes Dev. 2015. PMID: 25792598 Free PMC article. - Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation.
Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, Leonardi L, Tirone F, Grignani F. Passeri D, et al. Mol Cell Biol. 2006 Jul;26(13):5023-32. doi: 10.1128/MCB.01360-05. Mol Cell Biol. 2006. PMID: 16782888 Free PMC article.
References
- Beato, M., P. Herrlich, and G. Schütz. 1995. Steroid hormone receptors: many actors in search of a plot. Cell 83:851-857. - PubMed
- Chen, D., S.-M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. - PubMed
- Chen, D., H. Ma, H. Hong, S. S. Koh, S.-M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. - PubMed
- Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG00093/AG/NIA NIH HHS/United States
- GM61355/GM/NIGMS NIH HHS/United States
- R01 DK055274/DK/NIDDK NIH HHS/United States
- DK55274/DK/NIDDK NIH HHS/United States
- R37 DK055274/DK/NIDDK NIH HHS/United States
- T32 AG000093/AG/NIA NIH HHS/United States
- R01 GM061355/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous