Nr-CAM is a target gene of the beta-catenin/LEF-1 pathway in melanoma and colon cancer and its expression enhances motility and confers tumorigenesis - PubMed (original) (raw)
Nr-CAM is a target gene of the beta-catenin/LEF-1 pathway in melanoma and colon cancer and its expression enhances motility and confers tumorigenesis
Maralice E Conacci-Sorrell et al. Genes Dev. 2002.
Abstract
beta-catenin and plakoglobin (gamma-catenin) are homologous molecules involved in cell adhesion, linking cadherin receptors to the cytoskeleton. beta-catenin is also a key component of the Wnt pathway by being a coactivator of LEF/TCF transcription factors. To identify novel target genes induced by beta-catenin and/or plakoglobin, DNA microarray analysis was carried out with RNA from cells overexpressing either protein. This analysis revealed that Nr-CAM is the gene most extensively induced by both catenins. Overexpression of either beta-catenin or plakoglobin induced Nr-CAM in a variety of cell types and the LEF/TCF binding sites in the Nr-CAM promoter were required for its activation by catenins. Retroviral transduction of Nr-CAM into NIH3T3 cells stimulated cell growth, enhanced motility, induced transformation, and produced rapidly growing tumors in nude mice. Nr-CAM and LEF-1 expression was elevated in human colon cancer tissue and cell lines and in human malignant melanoma cell lines but not in melanocytes or normal colon tissue. Dominant negative LEF-1 decreased Nr-CAM expression and antibodies to Nr-CAM inhibited the motility of B16 melanoma cells. The results indicate that induction of Nr-CAM transcription by beta-catenin or plakoglobin plays a role in melanoma and colon cancer tumorigenesis, probably by promoting cell growth and motility.
Figures
Figure 1
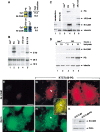
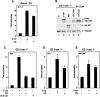
Induction of Nr-CAM expression by β-catenin and plakoglobin. (A) RNA was prepared from a mixture of three independent human renal carcinoma cell clones each of: cells stably expressing plakoglobin (PG), ΔNβ-catenin, or the neorgene alone. The RNA was tested, after Cy3 (PG and β-cat)- and Cy5 (neor)-labeling (following RT), for the expression of genes using an Incyte DNA microarray containing 10,000 ESTs. Fold induction of Nr-CAM expression compared with the positive (Cy3) control is shown. The blue color indicates no hybridization; the yellow and red area of the spectrum represent strong hybridization. (B) Northern blot hybridization for Nr-CAM using renal carcinoma cells stably expressing the genes described in A. B16 F10 melanoma cells served as positive control for Nr-CAM expression. (C) Western blot analysis for Nr-CAM in the cell clones indicated in A. Mouse cerebellum served as positive control for Nr-CAM and untransfected renal carcinoma cells (KTCTL60) as negative control. (D) KTCTL60 cells stably expressing plakoglobin (PG) and control neorcells were treated for different times with sodium butyrate to enhance the expression of the transgene (PG) and the expression of plakoglobin and Nr-CAM was determined by Western blotting. The levels of 18S and 28S rRNA in B, vinculin in C, and tubulin in D were used as controls for gel loading. (E–J) The localization of Nr-CAM and ezrin in cells stably transfected with plakoglobin. Untransfected (KTCTL60; E,F) and cells stably transfected with plakoglobin (KTCTL60-PG; G–J) were immunostained with anti Nr-CAM antibodies (E,G,J) and doubly stained with an antibody for ezrin (F,H). (I) Merged image of ezrin and Nr-CAM staining. The cells in J were stained for Nr-CAM and the image focuses at the cell periphery close to the substrate. (K) Expression of ezrin and Nr-CAM was determined by Western blotting. Note colocalization of Nr-CAM with ezrin-containing cellular protrusions (arrowhead). Bar in E, 10 μm.
Figure 2
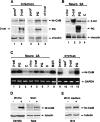
Retroviral transduction and transient transfection of β-catenin or plakoglobin induce the expression of Nr-CAM in different cell types. (A) KTCTL60 cells were infected with retroviruses expressing either a stable, HA-tagged β-catenin mutant S33Y (β-cat; lane 2), HA-tagged plakoglobin (PG; lane 4), or the puror gene (lanes 1,3). Cells stably expressing ΔNβ-catenin (lane 5) served as positive control. Expression of Nr-CAM and HA-tagged proteins was determined by Western blotting. The level of vinculin served as loading control. (B) Expression of the transfected genes in Neuro 2A cells was verified by Western blotting with antibodies to β-catenin, plakoglobin and vinculin. (C) Expression of Nr-CAM RNA in Neuro 2A cells transiently transfected either with a stable β-catenin mutant S33Y (β-cat; lanes 1,5), plakoglobin (PG; lanes 2,6), vinculin (vinc; lane 4), or untrasfected cells (c; lanes 3,7) was determined by RT-PCR and compared with that of B16 M2R melanoma cells (lane_8_), mouse brain tissue (lane 9), and KTCTL60 cells transfected with β-catenin (lane 11), plakoglobin (lane_12_), or the neor gene (lane 10). (D) Expression of Nr-CAM was also determined in C57MG and C57MG-Wnt7A and Rat2 and Rat2-Wnt-1 cells. (E) B16 M2R cells were incubated for 24 h with control medium and conditioned medium from Rat2–Wnt-1 cells and the level of Nr-CAM was determined by Western blotting.
Figure 3
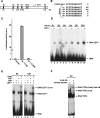
The Nr-CAM gene promoter contains several LEF-1 binding sites that can complex with LEF-1 and β-catenin. Schematic presentation of LEF-1 binding sites in the Nr-CAM gene promoter (A) and their nucleotide sequence compared to the consensus LEF/TCF binding sequence (B). (C) Activation of a synthetic 1.8-kb Nr-CAM promoter reporter containing the sense (S), but not the antisense (AS), sequence in 293T cells. (D) Binding of the different 32P-labeled LEF-1 sites in the Nr-CAM promoter to in vitro-transcribed–translated LEF-1 determined by the DNA mobility shift assay. The LEF-1 site of the cyclin D1 promoter (CD1) served as positive control. (E) Ternary complex formation by the LEF-1 binding site no. 4 (S4) of the Nr-CAM promoter with in vitro-translated LEF-1 and β-catenin was determined as in D. (F) Nuclear extracts from SW480 colon cancer cells displaying constitutive β-catenin-LEF/TCF signaling were incubated with32P-labeled LEF-1 site no. 4 (S4) and analyzed as in_D_, with (lane 2), or without (lane 1) incubation with anti β-catenin antibody. Note the supershift in the band representing the ternary complex in the presence of anti β-catenin antibody.
Figure 4
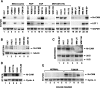
. Activation of the Nr-CAM promoter by β-catenin involves LEF-1 binding sites. (A) 293T cells were transfected with the Nr-CAM promoter reporter plasmid containing LEF-1 sites 1–4 (Nr 1–4) and with either wild-type β-catenin, the β-catenin mutant S33Y, ΔNβ-catenin, or plakoglobin (PG), and luciferase activity determined. (B) The levels of the proteins transfected in_A_ were determined by Western blotting. (C) The Nr-CAM reporter plasmid was cotransfected with a dominant negative TCF4 (ΔNTCF4), or with a plasmid expressing the cytoplasmic domain of E-cadherin (E-cad tail), and luciferase activity determined. (D) Schematic representation of Nr-CAM promoter constructs containing different LEF-1 sites. The activities of these Nr-CAM promoter constructs was determined in cells cotransfected with β-catenin (E), or with a chimeric dominant positive construct containing the C terminus of β-catenin and the DNA binding domain of LEF-1 (HMG-β-catenin; F). Note the weak ability of plakoglobin and inability of ΔNβ-catenin to activate the Nr-CAM promoter in 293T cells and the dependence on LEF-1 sites of Nr-CAM promoter activation by the dominant positive HMG-β-catenin chimera.
Figure 5
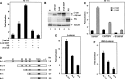
Plakoglobin can activate the Nr-CAM promoter in Neuro 2A cells and the cyclin D1 and Nr-CAM promoters in β-catenin−/− ES cells. (A) Neuro 2A cells were transfected with the Nr-CAM promoter plasmid together with either β-catenin (β-cat) or plakoglobin (PG) and luciferase activity was determined. (B,C) β-catenin−/− ES cells were transfected with the synthetic LEF/TCF reporter plasmid TOPFLASH (TOP) and either with β-catenin, plakoglobin or ΔNβ-catenin, and luciferase activity (C) and expression of the transfected proteins by Western blotting (B) were determined. (D,E) The abilities of β-catenin and plakoglobin to activate a cyclin D1 (CD1) promoter reporter (D) and the Nr-CAM promoter reporter (E) in β-catenin-null ES cells were determined. Note that plakoglobin can efficiently activate the Nr-CAM promoter in Neuro 2A cells, and the CD1 and Nr-CAM promoters and TOPFLASH in β-catenin−/− ES cells.
Figure 6
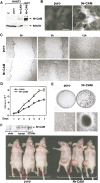
Retroviral infection of NIH3T3 cells with Nr-CAM enhances their capacity to migrate and close an artificial wound and confers tumorigenesis. (A) NIH3T3 cells were infected with a retrovirus coding for Nr-CAM and the _puro_r gene, or the _puro_r vector alone. Puromycin resistant cultures were isolated and expression of Nr-CAM compared by Western blotting to 293T cells transiently transfected with Nr-CAM. (B) Immunofluorescence detection of Nr-CAM localization in retrovirus infected NIH3T3 cells. (C) An artificial wound was introduced with a micropipette tip into confluent cultures of puror control and Nr-CAM infected NIH3T3 cells and the closure of this wound was followed for different times. (D) The proliferation of NIH3T3 cells expressing puror and Nr-CAM was compared. (E) Nr-CAM expressing 3T3 cells formed many large foci visualized at low magnification by Giemsa staining of the cells (top), or at higher magnification in live cultures (bottom). (F) Retrovirus-mediated transduction of Nr-CAM conferred the ability to form fast growing tumors when 106 cells were injected subcutaneously into nude mice. The picture was taken 14 d after injection and expression of Nr-CAM in tumor tissue and in normal skin and muscle was determined by Western blotting with anti Nr-CAM antibodies.
Figure 7
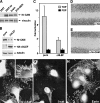
Melanoma cells express elevated Nr-CAM levels in membrane protrusions and nuclear β-catenin, and ΔNLEF reduces β-catenin/TCF activity and Nr-CAM expression in these cells. (A) Levels of Nr-CAM were determined by Western blotting in B16 M2R melanoma, brain tissue, 293T cells, and 293T cells transfected with Nr-CAM. (B) B16 M2R melanoma cells were infected with a retrovirus coding for dominant negative LEF-1 (ΔNLEF) and_puro_r and expression (in puror cultures) of Nr-CAM and HA-ΔNLEF was detected by Western blotting. (C) β-catenin-TCF-mediated transcription activity in purorcontrol and ΔNLEF cultures was determined by cotransfection of TOPFLASH (TOP) or the mutant FOPFLASH (FOP). The ratio between luciferase activity with TOP vs. FOP, after normalizing for transfection efficiency with co-transfected β-galactosidase, is presented. (D,E) The ability of B16 M2R melanoma to close a wound introduced in a monolayer was determined in cultures incubated for 14 h with a 1:20 dilution of polyclonal antibodies to HA (D) or Nr-CAM (E). (F) Endogenous Nr-CAM localized to membrane protrusions in B16 M2R cells that displayed nuclear β-catenin by double immunofluorescence staining (G). The localization of Nr-CAM was similar to that of ezrin, a marker of microvilli (H).
Figure 8
Expression of Nr-CAM is increased in human malignant melanoma and colon carcinoma tissue and cell lines. (A) Expression of Nr-CAM, LEF-1, and GAPDH, which served as control, was determined by RT-PCR in human melanocytes and human melanoma cell lines from different sources and stages of melanoma development, including the radial growth phase (RGP), vertical growth phase (VGP), and different metastatic loci (METASTATIC; see Materials and Methods). The level of Nr-CAM was also determined in some of these cell lines by Western (B) or Northern (C) blotting. (D) Nr-CAM levels in human colon cancer cell lines and (E) colon carcinoma and normal colon tissue samples was determined by RT-PCR using cyclophilin A as loading control. Note that whereas primary melanocytes and melanoma from the radial growth phase display almost no Nr-CAM expression, several metastatic melanoma cell lines show high levels of both Nr-CAM and LEF-1 expression. Also, note that although Nr-CAM was detected in all colon tumor tissue samples and cell lines, it was not detected in normal colon tissue.
Similar articles
- Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin.
Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, Ben-Ze'ev A. Simcha I, et al. J Cell Biol. 1998 Jun 15;141(6):1433-48. doi: 10.1083/jcb.141.6.1433. J Cell Biol. 1998. PMID: 9628899 Free PMC article. - Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter.
Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. Filali M, et al. J Biol Chem. 2002 Sep 6;277(36):33398-410. doi: 10.1074/jbc.M107977200. Epub 2002 Jun 6. J Biol Chem. 2002. PMID: 12052822 - Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells.
Sekiya T, Adachi S, Kohu K, Yamada T, Higuchi O, Furukawa Y, Nakamura Y, Nakamura T, Tashiro K, Kuhara S, Ohwada S, Akiyama T. Sekiya T, et al. J Biol Chem. 2004 Feb 20;279(8):6840-6. doi: 10.1074/jbc.M310876200. Epub 2003 Dec 1. J Biol Chem. 2004. PMID: 14660579 - Differential molecular interactions of beta-catenin and plakoglobin in adhesion, signaling and cancer.
Ben-Ze'ev A, Geiger B. Ben-Ze'ev A, et al. Curr Opin Cell Biol. 1998 Oct;10(5):629-39. doi: 10.1016/s0955-0674(98)80039-2. Curr Opin Cell Biol. 1998. PMID: 9818174 Review. - Plakoglobin and beta-catenin: protein interactions, regulation and biological roles.
Zhurinsky J, Shtutman M, Ben-Ze'ev A. Zhurinsky J, et al. J Cell Sci. 2000 Sep;113 ( Pt 18):3127-39. doi: 10.1242/jcs.113.18.3127. J Cell Sci. 2000. PMID: 10954412 Review.
Cited by
- Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling.
Heuberger J, Birchmeier W. Heuberger J, et al. Cold Spring Harb Perspect Biol. 2010 Feb;2(2):a002915. doi: 10.1101/cshperspect.a002915. Cold Spring Harb Perspect Biol. 2010. PMID: 20182623 Free PMC article. Review. - Loss of Function of the Neural Cell Adhesion Molecule NrCAM Regulates Differentiation, Proliferation and Neurogenesis in Early Postnatal Hypothalamic Tanycytes.
Moore A, Chinnaiya K, Kim DW, Brown S, Stewart I, Robins S, Dowsett GKC, Muir C, Travaglio M, Lewis JE, Ebling F, Blackshaw S, Furley A, Placzek M. Moore A, et al. Front Neurosci. 2022 Apr 7;16:832961. doi: 10.3389/fnins.2022.832961. eCollection 2022. Front Neurosci. 2022. PMID: 35464310 Free PMC article. - Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a.
Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Castelo-Branco G, et al. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12747-52. doi: 10.1073/pnas.1534900100. Epub 2003 Oct 13. Proc Natl Acad Sci U S A. 2003. PMID: 14557550 Free PMC article. - FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development.
Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. Chamorro MN, et al. EMBO J. 2005 Jan 12;24(1):73-84. doi: 10.1038/sj.emboj.7600460. Epub 2004 Dec 9. EMBO J. 2005. PMID: 15592430 Free PMC article. - Clusterin, a gene enriched in intestinal stem cells, is required for L1-mediated colon cancer metastasis.
Shapiro B, Tocci P, Haase G, Gavert N, Ben-Ze'ev A. Shapiro B, et al. Oncotarget. 2015 Oct 27;6(33):34389-401. doi: 10.18632/oncotarget.5360. Oncotarget. 2015. PMID: 26399194 Free PMC article.
References
- Aitkenhead M, Wang S-H, Nakatsu M, Mestas J, Heard C, Hughes C. Identification of endothelial cell genes expressed in an in vitro model of angiogenesis: induction of ESM-1 big-h3, and NrCAM. Microvascular Res. 2002;63:159–171. - PubMed
- Ben-Ze'ev A, Geiger B. Differential molecular interactions of β-catenin an plakoglobin in adhesion, signaling and cancer. Curr Opin Cell Biol. 1998;10:629–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous