Two classes of short interfering RNA in RNA silencing - PubMed (original) (raw)
Two classes of short interfering RNA in RNA silencing
Andrew Hamilton et al. EMBO J. 2002.
Erratum in
- Two classes of short interfering RNA in RNA silencing.
Hamilton A, Voinnet O, Chappell L, Baulcombe D. Hamilton A, et al. EMBO J. 2015 Oct 14;34(20):2590. doi: 10.15252/embj.201570050. Epub 2015 Aug 19. EMBO J. 2015. PMID: 26291654 Free PMC article. No abstract available.
Abstract
RNA silencing is a eukaryotic genome defence system that involves processing of double-stranded RNA (dsRNA) into 21-26 nt, short interfering RNA (siRNA). The siRNA mediates suppression of genes corresponding to the dsRNA through targeted RNA degradation. In some plant systems there are additional silencing processes, involving systemic spread of silencing and RNA-directed methylation/transcriptional suppression of homologous genomic DNA. We show here that siRNAs produced in plants from a green fluorescent protein (GFP) transgene are in short (21-22 nt) and long (24-26 nt) size classes, whereas those from endogenous retroelements are only in the long class. Viral suppressors of RNA silencing and mutations in Arabidopsis indicate that these classes of siRNA have different roles. The long siRNA is dispensable for sequence-specific mRNA degradation, but correlates with systemic silencing and methylation of homologous DNA. Conversely, the short siRNA class correlates with mRNA degradation but not with systemic signalling or methylation. These findings reveal an unexpected level of complexity in the RNA silencing pathway in plants that may also apply in animals.
Figures
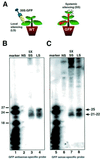
Fig. 1. Differential accumulation of long and short siRNA in local and systemic GFP silencing. (A) Local silencing (LS) of GFP (left) was induced in leaves of wild-type (WT) N.benthamiana by agro-infiltration of the 35S–GFP construct. Local silencing also occurs in GFP transgenic N.benthamiana (right) and precedes systemic silencing (SS), in which transgene expression is suppressed in new, emerging, non-infiltrated tissues. (B–C) Low molecular weight RNA was hybridized with GFP antisense-specific (B) or sense-specific (C) probes. Lanes ‘SS’ were from systemically silenced upper leaves of GFP transgenic N.benthamiana. ‘5X’ indicates that the amount of RNA loaded in lanes 3 and 7 was 5-fold higher than the amount loaded in lanes 4 and 8. The samples in lanes ‘LS’ were from leaves of wild-type N.benthamiana exhibiting local silencing following infiltration with the 35S–GFP strain of A.tumefaciens. Control samples (lanes ‘NS’) were from non-infiltrated leaves of wild-type N.benthamiana. Both GFP siRNA classes accumulate to similar, high levels in GFP transgenic and wild-type plants (data not shown).
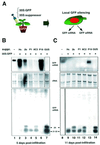
Fig. 2. The effects of viral suppressors on local RNA silencing. (A) Local silencing was induced in leaves of GFP transgenic N.benthamiana by infiltration of the 35S–GFP strain of Agrobacterium together with a second strain designed to express a viral suppressor of silencing. The onset of local silencing was then monitored and samples were collected for GFP mRNA and siRNA analysis. RNA was extracted from the infiltrated leaves after 5 (B) and 11 days (C). GFP mRNA was detected by hybridization with 32P-labelled GFP cDNA. GFP siRNA was detected by hybridization with labelled GFP sense RNA. The length of the siRNA is indicated as 21–22 nt (short class) and 25 nt (long class). Lanes 2–6 and 9–12 are from plants in which the GFP A.tumefaciens was mixed 1:1 with a second culture expressing a viral suppressor. The suppressors were Hc-Pro (HC, lanes 2 and 9), 2b (lanes 3 and 10), P1 (lanes 4 and 11), AC2 (lanes 5 and 12) and P19 (lanes 6 and 13). Lanes 7 and 14 are from plants in which the second A.tumefaciens culture carried the 35S–GUS transgene construct. Lanes 1 and 8 correspond to non-infiltrated control leaves.
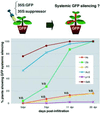
Fig. 3. The effects of viral suppressors on systemic RNA silencing. GFP transgenic N.benthamiana plants were infiltrated with mixed A.tumefaciens cultures as described in Figure 2, and were monitored for the onset of systemic silencing at 5, 7, 11 and 20 d.p.i. For each experimental treatment, a total of 30 individual seedlings (three separate experiments with 10 plants each) were tested. Abbreviations for the viral suppressors are as shown in the legend to Figure 2. V.O., veins only: this indicates that systemic silencing was incomplete and limited to the veins of a few leaves.
Fig. 4. The effects of PVX P25 on siRNA accumulation and systemic RNA silencing. (A) PVX–GFP has been described previously (Ruiz et al., 1998; Voinnet et al., 2000). Expression of the inserts in the PVX vector is controlled by a duplicated coat protein (CP) promoter, as indicated in blue. The replicase ORF is essential for viral replication; the 25, 12 and 8 kDa proteins are all strictly required for viral cell-to-cell movement and are collectively referred to as triple gene block (TGB) proteins. The CP is essential for encapsidation as well as cell-to-cell and systemic movement. PVX–GFP–ΔCP carries a deletion span ning the entire CP ORF. PVX–GFP–ΔTGB–ΔCP is based on PVX–GFP–ΔCP and carries a deletion spanning the entire TGB (Voinnet et al., 2000). These PVX–GFP mutants were inserted between the 35S promoter and terminator of the pBin61 T-DNA, and viral inocula were provided to plants by _Agrobacterium_-mediated transient expression of the above-mentioned T-DNAs. (B) Four-week-old seedlings of GFP transgenic N.benthamiana were inoculated with PVX–GFP-based replicons. After 5 days, RNA was extracted from the inoculated leaf and antisense GFP siRNA was assayed as described in Figure 2. The onset and progression of systemic GFP silencing was monitored in 10 plants for each inoculum and the number of plants showing systemic silencing is indicated. The asterisk at the bottom of lane 2 indicates partial systemic silencing that was only restricted to a few leaf veins in this particular plant. The viral inocula were either PVX–GFP–ΔTGB–ΔCP (lanes 1, 3 and 4) or PVX–GFP–ΔCP (lane 2). The 35S–P25–ΔATG (lane 3) and 35S–P25 (lane 4) constructs were transiently co-expressed in the inoculated tissue, as described in Figure 2.
Fig. 5. Retroelement siRNA in Nicotiana sp. (A) Low molecular weight RNA from N.benthamiana (lanes ‘N.b.’); 5-week-old N.tabacum var. Petite Havana (lanes ‘N.t.’) and 6-week-old A.thaliana landrace C24 (lanes ‘A.t.’) were hybridized with 32P-labelled Tnt1 LTR sense or antisense RNA. Samples in lanes ‘S’ and ‘NS’ were as described in Figure 1 and the siRNA was detected by hybridization with a GFP antisense-specific probe. (B) RNA was extracted from the leaves of 1-, 5- and 9-week-old N.tabacum var. Petite Havana. Low molecular weight RNA was hybridized with a 32P-labelled sense or antisense RNA corresponding to a cloned TS SINE. The two panels show hybridization to the same filter stripped of probe between hybridizations. (C) Leaves of N.benthamiana were inoculated with TRV:00 or TRV:TS and total RNA was extracted from the inoculated leaves after 2, 4 and 7 days. RNA was hybridized with a 32P-labelled, 617-bp DNA fragment of TRV RNA2 generated by _Eco_RI digestion of vector pTV00. This fragment corresponds to the 3′ UTR of RNA2 and thus cross-hybridizes with RNA1. The multiple RNA species hybridizing to the probe are genomic and subgenomic RNAs that are typically associated with TRV infection (Ratcliff et al., 2001). The size shift of the TRV:TS RNA species (lanes 7–9) confirms detection of the recombinant virus.
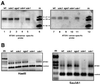
Fig. 6. Retroelement siRNA and DNA methylation in Arabidopsis. (A) RNA was extracted from wild-type and silencing-defective mutants of 6-week-old A.thaliana (landrace C24) seedlings. The mutations were at the sde1/sgs2, sgs3, sde3 and sde4 loci (Dalmay et al., 2000, 2001; Mourrain et al., 2000). AtSN1 siRNA was detected by hybridization with sense RNA transcribed from the cloned AtSN1 sequence. After probe stripping, the filter was reprobed with antisense RNA from the cloned AtSN1 sequence. (B) DNA was extracted from the lines of A.thaliana as in (A) and digested with the methylation-sensitive enzymes _Hae_III or _Sau_3AI. Similar extent of digestion was confirmed by electrophoresis and ethidium staining of digested DNA. Each sample of digested DNA was used as template for PCR with AtSN-specific primers and primers that would amplify an unrelated sequence (to control for variations in efficiency of individual PCR). The AtSN primers would only amplify AtSN DNA if it had remained undigested.
Similar articles
- Induction and suppression of RNA silencing by an animal virus.
Li H, Li WX, Ding SW. Li H, et al. Science. 2002 May 17;296(5571):1319-21. doi: 10.1126/science.1070948. Science. 2002. PMID: 12016316 - Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum.
Kościańska E, Kalantidis K, Wypijewski K, Sadowski J, Tabler M. Kościańska E, et al. Plant Mol Biol. 2005 Nov;59(4):647-61. doi: 10.1007/s11103-005-0668-x. Plant Mol Biol. 2005. PMID: 16244913 - Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing.
Vogt U, Pélissier T, Pütz A, Razvi F, Fischer R, Wassenegger M. Vogt U, et al. Plant J. 2004 Apr;38(1):107-18. doi: 10.1111/j.1365-313X.2004.02029.x. Plant J. 2004. PMID: 15053764 - The long and short of siRNAs.
Timmons L. Timmons L. Mol Cell. 2002 Sep;10(3):435-7. doi: 10.1016/s1097-2765(02)00657-3. Mol Cell. 2002. PMID: 12408811 Review. - Gene silencing: double-stranded RNA mediated mRNA degradation and gene inactivation.
Tang W, Luo XY, Sanmuels V. Tang W, et al. Cell Res. 2001 Sep;11(3):181-6. doi: 10.1038/sj.cr.7290084. Cell Res. 2001. PMID: 11642402 Review.
Cited by
- Protein P5 of pear chlorotic leaf spot-associated virus is a pathogenic factor that suppresses RNA silencing and enhances virus movement.
Ren Q, Zhang Z, Zhang Y, Zhang Y, Gao Y, Zhang H, Wang X, Wang G, Hong N. Ren Q, et al. Mol Plant Pathol. 2024 Oct;25(10):e70015. doi: 10.1111/mpp.70015. Mol Plant Pathol. 2024. PMID: 39412447 Free PMC article. - Regulatory mechanism of heat-active retrotransposons by the SET domain protein SUVH2.
Niu X, Ge Z, Ito H. Niu X, et al. Front Plant Sci. 2024 Feb 8;15:1355626. doi: 10.3389/fpls.2024.1355626. eCollection 2024. Front Plant Sci. 2024. PMID: 38390294 Free PMC article. - P4 protein of an Indian isolate of rice tungro bacilliform virus modulates gene silencing.
Naresh M, Purkayastha A, Dasgupta I. Naresh M, et al. Virus Genes. 2024 Feb;60(1):55-64. doi: 10.1007/s11262-023-02039-2. Epub 2023 Dec 6. Virus Genes. 2024. PMID: 38055154 - Conserved and non-conserved RNA-target modules in plants: lessons for a better understanding of Marchantia development.
Pietrykowska H, Alisha A, Aggarwal B, Watanabe Y, Ohtani M, Jarmolowski A, Sierocka I, Szweykowska-Kulinska Z. Pietrykowska H, et al. Plant Mol Biol. 2023 Nov;113(4-5):121-142. doi: 10.1007/s11103-023-01392-y. Epub 2023 Nov 22. Plant Mol Biol. 2023. PMID: 37991688 Free PMC article. Review. - Trans-grafting plum pox virus resistance from transgenic plum rootstocks to apricot scions.
Alburquerque N, Pérez-Caselles C, Faize L, Ilardi V, Burgos L. Alburquerque N, et al. Front Plant Sci. 2023 Sep 27;14:1216217. doi: 10.3389/fpls.2023.1216217. eCollection 2023. Front Plant Sci. 2023. PMID: 37828929 Free PMC article.
References
- Anandalakshmi R., Marathe,R., Ge,X., Herr,J.M., Mau,C., Mallory,A., Pruss,G., Bowman,L. and Vance,V.B. (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science, 290, 142–144. - PubMed
- Aravin A.A., Naumova,N.M., Tulin,A.V., Vagin,V.V., Rozovsky,Y.M. and Gvozdev,V.A. (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol., 11, 1017–1027. - PubMed
- Bendahmane A., Querci,M., Kanyuka,K. and Baulcombe,D.C. (2000) Agrobacterium transient expression system as a tool for isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J., 21, 73–81. - PubMed
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials