A comparison of profile hidden Markov model procedures for remote homology detection - PubMed (original) (raw)
Comparative Study
. 2002 Oct 1;30(19):4321-8.
doi: 10.1093/nar/gkf544.
Affiliations
- PMID: 12364612
- PMCID: PMC140544
- DOI: 10.1093/nar/gkf544
Comparative Study
A comparison of profile hidden Markov model procedures for remote homology detection
Martin Madera et al. Nucleic Acids Res. 2002.
Abstract
Profile hidden Markov models (HMMs) are amongst the most successful procedures for detecting remote homology between proteins. There are two popular profile HMM programs, HMMER and SAM. Little is known about their performance relative to each other and to the recently improved version of PSI-BLAST. Here we compare the two programs to each other and to non-HMM methods, to determine their relative performance and the features that are important for their success. The quality of the multiple sequence alignments used to build models was the most important factor affecting the overall performance of profile HMMs. The SAM T99 procedure is needed to produce high quality alignments automatically, and the lack of an equivalent component in HMMER makes it less complete as a package. Using the default options and parameters as would be expected of an inexpert user, it was found that from identical alignments SAM consistently produces better models than HMMER and that the relative performance of the model-scoring components varies. On average, HMMER was found to be between one and three times faster than SAM when searching databases larger than 2000 sequences, SAM being faster on smaller ones. Both methods were shown to have effective low complexity and repeat sequence masking using their null models, and the accuracy of their E-values was comparable. It was found that the SAM T99 iterative database search procedure performs better than the most recent version of PSI-BLAST, but that scoring of PSI-BLAST profiles is more than 30 times faster than scoring of SAM models.
Figures
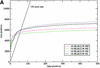
Figure 1
Sensitivity plots for the SCOP all-against-all (test 2). The input alignments were: (A) the results of a WU-BLAST search of nrdb90 aligned with ClustalW; (B) T99 alignments realigned with ClustalW. In both figures, HH and SS are the default procedures for HMMER and SAM, respectively, HS indicates a HMMER model converted to the SAM format and scored by SAM, and vice versa for SH.
Figure 1
Sensitivity plots for the SCOP all-against-all (test 2). The input alignments were: (A) the results of a WU-BLAST search of nrdb90 aligned with ClustalW; (B) T99 alignments realigned with ClustalW. In both figures, HH and SS are the default procedures for HMMER and SAM, respectively, HS indicates a HMMER model converted to the SAM format and scored by SAM, and vice versa for SH.
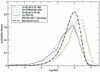
Figure 2
Distribution of E-values E of first false positives in test 2. The probability density is with respect to the log10(E) x_-axis. The experimental curves are smoothed (each model was added as a Gaussian of standard deviation 0.1 and area 2873–1), the theoretical curve is ln(10) E exp(–_E). 1S-BL&CLW is the result of a WU-BLAST search of nrdb90 aligned with ClustalW, 1S-T99 the alignment produced by the T99 procedure; HH and SS are the default procedures for HMMER and SAM, respectively.
Figure 3
Sensitivity plots for the SCOP all-against-all. SCOP version 1.50 was used, filtered down to 2873 sequences of less than 40% sequence identity, with a total of 36 612 possible true pairwise relationships. See the text for further details.
Similar articles
- Improved profile HMM performance by assessment of critical algorithmic features in SAM and HMMER.
Wistrand M, Sonnhammer EL. Wistrand M, et al. BMC Bioinformatics. 2005 Apr 15;6:99. doi: 10.1186/1471-2105-6-99. BMC Bioinformatics. 2005. PMID: 15831105 Free PMC article. - Protein homology detection by HMM-HMM comparison.
Söding J. Söding J. Bioinformatics. 2005 Apr 1;21(7):951-60. doi: 10.1093/bioinformatics/bti125. Epub 2004 Nov 5. Bioinformatics. 2005. PMID: 15531603 - A comparison of scoring functions for protein sequence profile alignment.
Edgar RC, Sjölander K. Edgar RC, et al. Bioinformatics. 2004 May 22;20(8):1301-8. doi: 10.1093/bioinformatics/bth090. Epub 2004 Feb 12. Bioinformatics. 2004. PMID: 14962936 - Profile hidden Markov models.
Eddy SR. Eddy SR. Bioinformatics. 1998;14(9):755-63. doi: 10.1093/bioinformatics/14.9.755. Bioinformatics. 1998. PMID: 9918945 Review. - Sequence comparison and protein structure prediction.
Dunbrack RL Jr. Dunbrack RL Jr. Curr Opin Struct Biol. 2006 Jun;16(3):374-84. doi: 10.1016/j.sbi.2006.05.006. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713709 Review.
Cited by
- Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice.
Sharma R, Agarwal P, Ray S, Deveshwar P, Sharma P, Sharma N, Nijhawan A, Jain M, Singh AK, Singh VP, Khurana JP, Tyagi AK, Kapoor S. Sharma R, et al. Funct Integr Genomics. 2012 Jun;12(2):229-48. doi: 10.1007/s10142-012-0274-3. Epub 2012 Mar 31. Funct Integr Genomics. 2012. PMID: 22466020 - The identification of complete domains within protein sequences using accurate E-values for semi-global alignment.
Kann MG, Sheetlin SL, Park Y, Bryant SH, Spouge JL. Kann MG, et al. Nucleic Acids Res. 2007;35(14):4678-85. doi: 10.1093/nar/gkm414. Epub 2007 Jun 27. Nucleic Acids Res. 2007. PMID: 17596268 Free PMC article. - A sequence sub-sampling algorithm increases the power to detect distant homologues.
Johnston CR, Shields DC. Johnston CR, et al. Nucleic Acids Res. 2005 Jul 8;33(12):3772-8. doi: 10.1093/nar/gki687. Print 2005. Nucleic Acids Res. 2005. PMID: 16006623 Free PMC article. - MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress.
Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. Arora R, et al. BMC Genomics. 2007 Jul 18;8:242. doi: 10.1186/1471-2164-8-242. BMC Genomics. 2007. PMID: 17640358 Free PMC article. - Bacterial-type ferroxidase tunes iron-dependent phosphate sensing during Arabidopsis root development.
Naumann C, Heisters M, Brandt W, Janitza P, Alfs C, Tang N, Toto Nienguesso A, Ziegler J, Imre R, Mechtler K, Dagdas Y, Hoehenwarter W, Sawers G, Quint M, Abel S. Naumann C, et al. Curr Biol. 2022 May 23;32(10):2189-2205.e6. doi: 10.1016/j.cub.2022.04.005. Epub 2022 Apr 25. Curr Biol. 2022. PMID: 35472311 Free PMC article.
References
- Park J., Karplus,K., Barrett,C., Hughey,R., Haussler,D., Hubbard,T. and Chothia,C. (1998) Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol., 284, 1201–1210. - PubMed
- Krogh A., Brown,M., Mian,S., Sjolander,K. and Haussler,D. (1994) Hidden Markov models in computational biology. J. Mol. Biol., 235, 1501–1531. - PubMed
- Eddy S.R. (1995) Hidden Markov models. Curr. Opin. Struct. Biol., 6, 361–365. - PubMed
- Lindahl E. and Elofsson,A. (2000) Identification of related proteins on family, superfamily and fold level. J. Mol. Biol., 295, 613–625. - PubMed
- Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) A new approach to protein fold recognition. Nature, 358, 86–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials