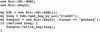The Bioperl toolkit: Perl modules for the life sciences - PubMed (original) (raw)
. 2002 Oct;12(10):1611-8.
doi: 10.1101/gr.361602.
David Block, Kris Boulez, Steven E Brenner, Stephen A Chervitz, Chris Dagdigian, Georg Fuellen, James G R Gilbert, Ian Korf, Hilmar Lapp, Heikki Lehväslaiho, Chad Matsalla, Chris J Mungall, Brian I Osborne, Matthew R Pocock, Peter Schattner, Martin Senger, Lincoln D Stein, Elia Stupka, Mark D Wilkinson, Ewan Birney
Affiliations
- PMID: 12368254
- PMCID: PMC187536
- DOI: 10.1101/gr.361602
The Bioperl toolkit: Perl modules for the life sciences
Jason E Stajich et al. Genome Res. 2002 Oct.
Abstract
The Bioperl project is an international open-source collaboration of biologists, bioinformaticians, and computer scientists that has evolved over the past 7 yr into the most comprehensive library of Perl modules available for managing and manipulating life-science information. Bioperl provides an easy-to-use, stable, and consistent programming interface for bioinformatics application programmers. The Bioperl modules have been successfully and repeatedly used to reduce otherwise complex tasks to only a few lines of code. The Bioperl object model has been proven to be flexible enough to support enterprise-level applications such as EnsEMBL, while maintaining an easy learning curve for novice Perl programmers. Bioperl is capable of executing analyses and processing results from programs such as BLAST, ClustalW, or the EMBOSS suite. Interoperation with modules written in Python and Java is supported through the evolving BioCORBA bridge. Bioperl provides access to data stores such as GenBank and SwissProt via a flexible series of sequence input/output modules, and to the emerging common sequence data storage format of the Open Bioinformatics Database Access project. This study describes the overall architecture of the toolkit, the problem domains that it addresses, and gives specific examples of how the toolkit can be used to solve common life-sciences problems. We conclude with a discussion of how the open-source nature of the project has contributed to the development effort.
Figures
Figure 1
Rendering a sequence graphically with Bio∷Graphics. This image represents a 20-Kb segment of the C. elegans genome containing annotated genes, a cross-species alignment (C. elegans to C. briggsae), EST alignments, SNPs, PCR primer pairs, and a GC content histogram. The module's flexible glyph-based architecture allows the application programmer to adjust precisely how to display biological objects. Glyphs allow the programmer to define different symbols for different data types or data sources and each are drawn as a separate track in the image. The module is also suitable for illustrating the extent of protein domains, physical (clone) maps, and horizontal maps.
Figure 2
This figure shows a portion of the Bioperl object model including the interfaces (shown in italicized type) for sequences (PrimarySeqI, SeqI, RichSeqI) and their implementations PrimarySeq (general sequence), Seq (sequence with features), RichSeq (sequence with features and rich annotation), LargePrimarySeq (for sequences too large to be held in a program's memory), and LargeSeq (large sequences with features). Also included in the diagram is the sequence feature interface (SeqFeatureI) and its implementations Similarity (manage similarity information), FeaturePair (paired feature information), and SimilarityPair (paired similarity information such as a pair-wise alignment information). Additionally, the diagram shows the location objects that manage Simple (start, end, and strand information), Split (multiple start and end spots on a sequence such as a set of exons), and so-called Fuzzy locations (where start, end or span is not exact) for sequence features.
Figure 3
Retrieving a sequence from a remote database with Bio∷DB∷EMBL. This code retrieves an mRNA sequence in EMBL format from the EBI EMBL databank with the accession no. U14680 and writes the sequence out in GenBank format to the terminal. One could replace Bio∷DB∷EMBL with Bio∷DB∷GenBank and instead retrieve the sequence from NCBI just as easily, as the software can read and write both EMBL and GenBank formats and is able to connect to both services through the World Wide Web. The retrieved sequence can then be passed to Bio∷Graphics for graphical rendering, to the Bio∷SeqIO interface for writing to a file, or to the ODBA interfaces for storage in a relational database.
Figure 4
Report parsing with Bio∷SearchIO. This code parses a BLAST report from a file called report.bls and saves, in an array called @HitsToSave, only the hits that have High-scoring Segment Pairs (HSPs) meeting an e-value and length threshold. In this case, any hit with e-value >0.001 or length < 120 residues will be excluded. Once the array is built, the names of each of the hits that had a HSP that met the criteria are printed out. To parse a FASTA (Pearson and Lipman 1988) report file one simply changes the format specification from blast to fasta.
Similar articles
- BpWrapper: BioPerl-based sequence and tree utilities for rapid prototyping of bioinformatics pipelines.
Hernández Y, Bernstein R, Pagan P, Vargas L, McCaig W, Ramrattan G, Akther S, Larracuente A, Di L, Vieira FG, Qiu WG. Hernández Y, et al. BMC Bioinformatics. 2018 Mar 2;19(1):76. doi: 10.1186/s12859-018-2074-9. BMC Bioinformatics. 2018. PMID: 29499649 Free PMC article. - A suite of Perl modules for handling microarray data.
Morris JA, Gayther SA, Jacobs IJ, Jones C. Morris JA, et al. Bioinformatics. 2008 Apr 15;24(8):1102-3. doi: 10.1093/bioinformatics/btn085. Epub 2008 Mar 18. Bioinformatics. 2008. PMID: 18353790 - Wildfire: distributed, Grid-enabled workflow construction and execution.
Tang F, Chua CL, Ho LY, Lim YP, Issac P, Krishnan A. Tang F, et al. BMC Bioinformatics. 2005 Mar 24;6:69. doi: 10.1186/1471-2105-6-69. BMC Bioinformatics. 2005. PMID: 15788106 Free PMC article. - Workflow based framework for life science informatics.
Tiwari A, Sekhar AK. Tiwari A, et al. Comput Biol Chem. 2007 Oct;31(5-6):305-19. doi: 10.1016/j.compbiolchem.2007.08.009. Epub 2007 Aug 19. Comput Biol Chem. 2007. PMID: 17931570 Review. - A library of efficient bioinformatics algorithms.
Della Vedova G, Dondi R. Della Vedova G, et al. Appl Bioinformatics. 2003;2(2):117-21. Appl Bioinformatics. 2003. PMID: 15130828 Review.
Cited by
- Biosynthetic gene clusters from uncultivated soil bacteria of the Atacama Desert.
Andreani-Gerard CM, Cambiazo V, González M. Andreani-Gerard CM, et al. mSphere. 2024 Oct 29;9(10):e0019224. doi: 10.1128/msphere.00192-24. Epub 2024 Sep 17. mSphere. 2024. PMID: 39287428 Free PMC article. - Bioinformatics analysis of SH2D4A in glioblastoma multiforme to evaluate immune features and predict prognosis.
Yang T, Li C, Xu D, Quan R, Wang L, Ren Y, Zhang Z, Yu R. Yang T, et al. Transl Cancer Res. 2024 Aug 31;13(8):4242-4256. doi: 10.21037/tcr-23-2000. Epub 2024 Aug 23. Transl Cancer Res. 2024. PMID: 39262462 Free PMC article. - Impact of the chemical modification of tRNAs anticodon loop on the variability and evolution of codon usage in proteobacteria.
Delgado S, Armijo Á, Bravo V, Orellana O, Salazar JC, Katz A. Delgado S, et al. Front Microbiol. 2024 Aug 5;15:1412318. doi: 10.3389/fmicb.2024.1412318. eCollection 2024. Front Microbiol. 2024. PMID: 39161601 Free PMC article. - Genome-wide identification, characterization and expression analysis of the bZIP transcription factors in garlic (Allium sativum L.).
He S, Xu S, He Z, Hao X. He S, et al. Front Plant Sci. 2024 Aug 1;15:1391248. doi: 10.3389/fpls.2024.1391248. eCollection 2024. Front Plant Sci. 2024. PMID: 39148621 Free PMC article. - Ecological genomics in the Northern krill uncovers loci for local adaptation across ocean basins.
Unneberg P, Larsson M, Olsson A, Wallerman O, Petri A, Bunikis I, Vinnere Pettersson O, Papetti C, Gislason A, Glenner H, Cartes JE, Blanco-Bercial L, Eriksen E, Meyer B, Wallberg A. Unneberg P, et al. Nat Commun. 2024 Aug 1;15(1):6297. doi: 10.1038/s41467-024-50239-7. Nat Commun. 2024. PMID: 39090106 Free PMC article.
References
- Achard F, Vaysseix G, Barillot E. XML, Bioinformatics, and data integration. Bioinformatics. 2001;17:115–125. - PubMed
- Beck K. Extreme programming examined: Embrace change. Reading, MA: Addison Wesley; 1999.
- Burge C, Karlin S. Prediction of complete gene stuctures in human genomic DNA. J Mol Biol. 1997;268:78–94. - PubMed
- Chervitz SA, Fuellen G, Dagdigian C, Brenner SE, Birney E, Korf I. Bioperl: Standard perl modules for bioinformatics. Bio Informatics Technology and Systems (BITS) 1998. http://www.bitsjournal.com/bioperl.html , http://www.bitsjournal.com/bioperl.html. .
Publication types
MeSH terms
Grants and funding
- T32 GM007754/GM/NIGMS NIH HHS/United States
- U41 HG000739/HG/NHGRI NIH HHS/United States
- 1 K32 HG00056/HG/NHGRI NIH HHS/United States
- P41 HG002223/HG/NHGRI NIH HHS/United States
- K22 HG000064/HG/NHGRI NIH HHS/United States
- K22 HG000056/HG/NHGRI NIH HHS/United States
- K22 HG-00064-01/HG/NHGRI NIH HHS/United States
- P41 HG000739/HG/NHGRI NIH HHS/United States
- P41HG02223/HG/NHGRI NIH HHS/United States
- HG00739/HG/NHGRI NIH HHS/United States
- T32 GM07754-22/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous