MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells - PubMed (original) (raw)
MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells
María C de Marco et al. J Cell Biol. 2002.
Abstract
Transcytosis is used alone (e.g., hepatoma HepG2 cells) or in combination with a direct pathway from the Golgi (e.g., epithelial MDCK cells) as an indirect route for targeting proteins to the apical surface. The raft-associated MAL protein is an essential element of the machinery for the direct route in MDCK cells. Herein, we present the functional characterization of MAL2, a member of the MAL protein family, in polarized HepG2 cells. MAL2 resided selectively in rafts and is predominantly distributed in a compartment localized beneath the subapical F-actin cytoskeleton. MAL2 greatly colocalized in subapical endosome structures with transcytosing molecules en route to the apical surface. Depletion of endogenous MAL2 drastically blocked transcytotic transport of exogenous polymeric immunoglobulin receptor and endogenous glycosylphosphatidylinositol-anchored protein CD59 to the apical membrane. MAL2 depletion did not affect the internalization of these molecules but produced their accumulation in perinuclear endosome elements that were accessible to transferrin. Normal transcytosis persisted in cells that expressed exogenous MAL2 designed to resist the depletion treatment. MAL2 is therefore essential for transcytosis in HepG2 cells.
Figures
Figure 1.
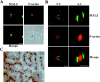
Characterization of human MAL2 with a newly generated monoclonal antibody. (A) Schematic model of the predicted structure of MAL2 with indication of the position of the consensus _N_-glycosylation site present in the molecule and the NH2-terminal peptide selected for the preparation of antibodies. (B) Expression of the MAL2 gene in different cell lines. Total RNA from the indicated cell lines was hybridized to DNA probes specific to MAL2, MAL, or β-actin. (C) Characterization of a novel mAb to MAL2. To assay the specificity of mAb 9D1, protein extracts from untransfected (U) or from transfected (T) COS-7 cells transiently expressing MAL2 tagged with the c-Myc 9E10 epitope were subjected to immunoblot analysis with either mAb 9D1 or the antitag mAb 9E10. Since COS-7 cells are negative for MAL2 gene expression (unpublished data), no reaction was observed with endogenous proteins of COS-7 cells. (D) mAb 9D1 detects endogenous MAL2. Membrane fractions from the indicated cell lines were subjected to immunoblot analysis with anti-MAL2 mAb 9D1. (E) Identification of endogenous MAL2 in lipid raft fractions in HepG2 cells. Cells were extracted with 1% Triton X-100 at 4°C and subjected to centrifugation to equilibrium in sucrose density gradients. Aliquots from each fraction were analyzed by immunoblotting with anti-MAL2 mAb 9D1, and antibodies to CD59, used as a raft marker, and to TfR, a transmembrane protein excluded from rafts. Fractions 1–4 represent the 40% sucrose layer and contain the bulk of cellular membranes and cytosolic proteins, whereas fractions 5–12 represent the 5–30% sucrose layer and contain the rafts.
Figure 2.
Distribution of MAL2 in polarized HepG2 cells and human liver. (A) HepG2 cells were double stained for MAL2 and F-actin and analyzed using a conventional fluorescence microscope. A phase–contrast image from the same field was taken to visualize the bile canaliculus (arrowhead). (B) HepG2 cells were double stained for MAL2 and F-actin and analyzed by confocal microscopy. Optical horizontal x,y and vertical x,z sections corresponding to planes in the middle of the cell or in the center of the bile canaliculus, respectively, are shown. (C) Liver tissue sections were subjected to immunohistochemical analysis with anti-MAL2 mAb 9D1 and counterstained with hematoxylin to visualize nuclei. Reactivity was found exclusively in the bile canaliculi, which appear as dots or lines depending on whether they were sectioned transversely (filled arrowheads) or longitudinally (open arrowheads) and absent from the sinusoidal membrane (asterisks). The inset shows at higher magnification a transversely sectioned bile canaliculus stained for MAL2. Bars, 5 μm.
Figure 3.
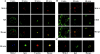
Double label immunofluorescence analysis of MAL2 and trancytosing molecules. (A) HepG2 cells stably expressing pIgR were incubated with IgA for 30 min at 4°C, washed, and placed at 37°C for the indicated times. Cells were then subjected to double label immunofluorescence analysis as indicated to detect MAL2 and IgA or were triply labeled to also detect F-actin. (B) HepG2 cells were incubated for 30 min at 4°C with anti-CD59 mAb, washed, and incubated at 37°C for the indicated times. Cells were then subjected to double label immunofluorescence analysis to detect MAL2 and CD59. (C) HepG2 cells stably expressing pIgR were incubated with IgA for 30 min at 4°C, washed, and incubated at 37°C for 60 min in the presence of 33 μM nocodazole. Cells were then washed, incubated for 120 min at 18°C in the absence of nocodazole, and subjected to double label immunofluorescence analysis to detect MAL2 and IgA. In all cases, the images presented are the composite projection of all the frames obtained by confocal microscopic analysis of the cells. Bars, 5 μm.
Figure 4.
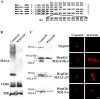
Depletion of endogenous MAL2 by transfection with an antisense phosphorothioate oligonucleotide. (A) The sequence of the antisense oligonucleotide used in MAL2 depletion experiments (MAL2/AS) and its alignment with wild-type MAL2 mRNA and the recombinant MAL2 mRNA species expressed in HepG2 cells are shown. Note that the changes introduced in the recombinant MAL2 transcripts prevent pairing with oligonucleotide MAL2/AS. The singly underlined residues correspond to sequences in the vector located immediately upstream of the inserted cDNA sequence. The doubly underlined residues in the coding sequence indicate nucleotides replaced by an equivalent triplet (MAL2-e), deleted (MAL2-ΔN) or added (MAL2-Myc). (B) HepG2 cells were transfected with control or MAL2/AS oligonucleotide and incubated at 37°C. After 72 h, cell extracts were subjected to immunoblot analysis with anti-MAL2 mAb 9D1 and with anti-CD59 and TfR antibodies. (C) Normal HepG2 cells or cells stably expressing the indicated exogenous products were transfected with control or MAL2/AS oligonucleotide and incubated at 37°C. After 72 h, cells were processed for immunoblot and immunofluorescence analyses with anti-MAL2 mAb, which recognizes both endogenous and exogenous MAL2 species. For simplicity, only the part of the blots corresponding to the unglycosylated MAL2 species is shown. Note that due to the deletion (MAL2-ΔN) or insertion (MAL2-Myc) of sequences the modified MAL2 proteins migrates faster or slower, respectively, than endogenous MAL2. Several independent cell clones (>3) from each type of cell transfectant were assayed and gave the same results. The immunofluorescence images correspond to the composite projection of all the frames obtained by confocal microscopic analysis of the cells. Bar, 5 μm.
Figure 5.
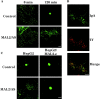
MAL2 depletion blocks transcytosis of both pIgR-IgA and CD59 in HepG2 cells. (A) HepG2 cells stably expressing pIgR were transfected with either control or MAL2/AS oligonucleotide, plated, and incubated at 37°C for 72 h. Cells were incubated in the presence of IgA for 60 min at 4°C and then were treated with 33 μM nocodazole for 60 min at 37°C. After removal of nocodazole (0 min), cells were incubated at 37°C for 120 min in the absence of the drug. Cells were stained with anti-IgA antibodies to determine the distribution of IgA. In parallel, mAb 9D1 was used to visualize the effect of the antisense oligonucleotide on MAL2 levels (unpublished data). One representative experiment out of ten performed is shown. (B) HepG2 cells in which IgA was accumulated in perinuclear endosomes by depletion of MAL2 were incubated with Tf for 10 min at 37°C, washed, and incubated for 10 min at 37°C to allow trafficking of internalized Tf to downstream compartments. Cells were then subjected to double label immunofluorescence analysis to detect IgA and Tf. (C) Normal HepG2 cells or HepG2 cells stably expressing MAL2-e were transfected with either control or MAL2/AS oligonucleotide, plated, and incubated at 37°C for 72 h. After binding of anti-CD59 mAb at 4°C, cells were incubated at 37°C for 60 min and fixed. After permeabilization, cells were stained with secondary antibodies to visualize the antibody bound CD59 complexes. One representative experiment out of six performed is shown. In all cases, the images presented are the composite projection of all the frames obtained by confocal microscopic analysis of the cells. Bars, 5 μm.
Similar articles
- Dynamics of MAL2 during glycosylphosphatidylinositol-anchored protein transcytotic transport to the apical surface of hepatoma HepG2 cells.
de Marco MC, Puertollano R, Martínez-Menárguez JA, Alonso MA. de Marco MC, et al. Traffic. 2006 Jan;7(1):61-73. doi: 10.1111/j.1600-0854.2005.00361.x. Traffic. 2006. PMID: 16445687 - Expression and distribution of MAL2, an essential element of the machinery for basolateral-to-apical transcytosis, in human thyroid epithelial cells.
Marazuela M, Martín-Belmonte F, García-López MA, Aranda JF, de Marco MC, Alonso MA. Marazuela M, et al. Endocrinology. 2004 Feb;145(2):1011-6. doi: 10.1210/en.2003-0652. Epub 2003 Oct 23. Endocrinology. 2004. PMID: 14576188 - Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue.
Marazuela M, Alonso MA. Marazuela M, et al. Histol Histopathol. 2004 Jul;19(3):925-33. doi: 10.14670/HH-19.925. Histol Histopathol. 2004. PMID: 15168355 Review. - The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2.
Madrid R, Aranda JF, Rodríguez-Fraticelli AE, Ventimiglia L, Andrés-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gómez S, Jiménez A, Martín-Belmonte F, Byrne JA, Alonso MA. Madrid R, et al. Dev Cell. 2010 May 18;18(5):814-27. doi: 10.1016/j.devcel.2010.04.001. Dev Cell. 2010. PMID: 20493814 - Research advances of MAL family members in tumorigenesis and tumor progression (Review).
Li M, Du Y, Zhang X, Zhou W. Li M, et al. Mol Med Rep. 2024 Apr;29(4):57. doi: 10.3892/mmr.2024.13181. Epub 2024 Feb 16. Mol Med Rep. 2024. PMID: 38362940 Free PMC article. Review.
Cited by
- Transcytosis in the development and morphogenesis of epithelial tissues.
Serra ND, Sundaram MV. Serra ND, et al. EMBO J. 2021 May 3;40(9):e106163. doi: 10.15252/embj.2020106163. Epub 2021 Apr 1. EMBO J. 2021. PMID: 33792936 Free PMC article. Review. - Resistance to Trastuzumab.
Vivekanandhan S, Knutson KL. Vivekanandhan S, et al. Cancers (Basel). 2022 Oct 19;14(20):5115. doi: 10.3390/cancers14205115. Cancers (Basel). 2022. PMID: 36291900 Free PMC article. Review. - Polarized sorting and trafficking in epithelial cells.
Cao X, Surma MA, Simons K. Cao X, et al. Cell Res. 2012 May;22(5):793-805. doi: 10.1038/cr.2012.64. Epub 2012 Apr 24. Cell Res. 2012. PMID: 22525333 Free PMC article. Review. - Cholesterol-sensitive modulation of transcytosis.
Leyt J, Melamed-Book N, Vaerman JP, Cohen S, Weiss AM, Aroeti B. Leyt J, et al. Mol Biol Cell. 2007 Jun;18(6):2057-71. doi: 10.1091/mbc.e06-08-0735. Epub 2007 Mar 28. Mol Biol Cell. 2007. PMID: 17392516 Free PMC article. - MAL2 mediates the formation of stable HER2 signaling complexes within lipid raft-rich membrane protrusions in breast cancer cells.
Jeong J, Shin JH, Li W, Hong JY, Lim J, Hwang JY, Chung JJ, Yan Q, Liu Y, Choi J, Wysolmerski J. Jeong J, et al. Cell Rep. 2021 Dec 28;37(13):110160. doi: 10.1016/j.celrep.2021.110160. Cell Rep. 2021. PMID: 34965434 Free PMC article.
References
- Brown, D.A., and J.K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68:533–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous