Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways - PubMed (original) (raw)
Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways
Luis Alfonso Martinez et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Single base pair mutations that alter the function of tumor suppressor genes and oncogenes occur frequently during oncogenesis. The guardian of the genome, p53, is inactivated by point mutation in more than 50% of human cancers. Synthetic small inhibiting RNAs (siRNAs) can suppress gene expression in mammalian cells, although their degree of selectivity might be compromised by an amplification mechanism. Here, we demonstrate that a single base difference in siRNAs discriminates between mutant and WT p53 in cells expressing both forms, resulting in the restoration of WT protein function. Therefore, siRNAs may be used to suppress expression of point-mutated genes and provide the basis for selective and personalized antitumor therapy.
Figures
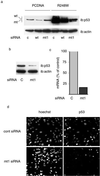
Fig 1.
Selective inhibition of p53 by siRNAs. (a) Sequences of the siRNAs used in this study; mutated nucleotides are indicated in bold. (b) H1299 cells were transfected with expression vectors for WT (WT) or R248W mutant (mt) p53, together with 400 ng of control (c), WT (WT), or mutant siRNA (mt1), as indicated. Cells were harvested 24 h later and analyzed by Western blotting (ib, immunoblot) with the indicated antibodies.
Fig 2.
Selectivity of mutant p53 inhibition. (a) Schematic model of a putative amplification process that would abolish the specificity of targeting point-mutated sequences: the antisense strand of the siRNA (arrow) is used as a primer by a putative RNA-dependent RNA polymerase for RNA strand elongation, resulting in a double-stranded 5′ portion of the target mRNA; this double-stranded sequence would in turn serve as a substrate for DICER which would result in siRNAs corresponding to the 5′ end of the target, which is common to both WT and mutant siRNAs. The square represents the single base mutation. (b and c) H1299 cells were transfected as in Fig. 1, with WT or R248W mutant (mt) expression vectors, alone or in combination, as indicated, along with the specified siRNAs, and analyzed by Western blotting 24 h later with the indicated antibodies.
Fig 3.
Inhibition of endogenous mutant p53. (a) Stable transfectants were established from U2OS cells (expressing WT p53) by using a mutant expression vector (R248W) or the corresponding empty vehicle vector (PCDNA). Cells were analyzed by Western blotting with the indicated antibodies. (b_–_d) Fibroblasts from a Li–Fraumeni patient were transfected with siRNAs and analyzed for p53 expression by Western blotting (b), real-time RT-PCR (c), or immunofluorescence (d).
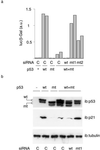
Fig 4.
Restoration of WT p53 function. (a) H1299 cells were transfected with both WT and mutant p53 vectors, siRNAs as indicated, and an mdm2-luciferase reporter construct along with a β-galactosidase construct as an internal control; cells were harvested 24 h later and extracts were assayed for luciferase activity. Transfections were performed in duplicate. Luciferase assays were performed in triplicate. Results are shown of two independent transfections from a representative experiment. (b) H1299 cells were transfected as in a and analyzed by Western blotting with the indicated antibodies.
Fig 5.
Immortalized Li–Fraumeni cells (MDAH087) were treated with control or mt1-siRNA as indicated. (a) Western blot analysis. (b) Terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) analysis (proportions of TUNEL-positive cells are shown in percentages).
Fig 6.
Restoration of p53 function in E6-expressing cells. (a) CasKi and SiHa cells that express HPV E6 protein were transfected with indicated siRNAs, treated with doxorubicin, or left untreated for the controls and then analyzed by Western blotting with the indicated antibodies. (b and c) SiHA cells treated as in a were analyzed for cell cycle. The result of a typical experiment is shown.
Similar articles
- Elevated expression of ribosomal protein genes L37, RPP-1, and S2 in the presence of mutant p53.
Loging WT, Reisman D. Loging WT, et al. Cancer Epidemiol Biomarkers Prev. 1999 Nov;8(11):1011-6. Cancer Epidemiol Biomarkers Prev. 1999. PMID: 10566557 - Death-associated protein kinase 1 promotes growth of p53-mutant cancers.
Zhao J, Zhao D, Poage GM, Mazumdar A, Zhang Y, Hill JL, Hartman ZC, Savage MI, Mills GB, Brown PH. Zhao J, et al. J Clin Invest. 2015 Jul 1;125(7):2707-20. doi: 10.1172/JCI70805. Epub 2015 Jun 15. J Clin Invest. 2015. PMID: 26075823 Free PMC article. - The p53 tumor suppressor gene and gene product.
Levine AJ. Levine AJ. Princess Takamatsu Symp. 1989;20:221-30. Princess Takamatsu Symp. 1989. PMID: 2488233 Review. - Induction of genetic instability by gain-of-function p53 cancer mutants.
Xu Y. Xu Y. Oncogene. 2008 Jun 5;27(25):3501-7. doi: 10.1038/sj.onc.1211023. Epub 2008 Jan 28. Oncogene. 2008. PMID: 18223686 Review.
Cited by
- Progress towards in vivo use of siRNAs.
Behlke MA. Behlke MA. Mol Ther. 2006 Apr;13(4):644-70. doi: 10.1016/j.ymthe.2006.01.001. Epub 2006 Feb 14. Mol Ther. 2006. PMID: 16481219 Free PMC article. Review. - RNA interference: from gene silencing to gene-specific therapeutics.
Leung RK, Whittaker PA. Leung RK, et al. Pharmacol Ther. 2005 Aug;107(2):222-39. doi: 10.1016/j.pharmthera.2005.03.004. Pharmacol Ther. 2005. PMID: 15908010 Free PMC article. Review. - Strand-specific 5'-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity.
Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Chen PY, et al. RNA. 2008 Feb;14(2):263-74. doi: 10.1261/rna.789808. Epub 2007 Dec 19. RNA. 2008. PMID: 18094121 Free PMC article. - Designing siRNA that distinguish between genes that differ by a single nucleotide.
Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Schwarz DS, et al. PLoS Genet. 2006 Sep 8;2(9):e140. doi: 10.1371/journal.pgen.0020140. Epub 2006 Jul 24. PLoS Genet. 2006. PMID: 16965178 Free PMC article. - Personalized cancer approach: using RNA interference technology.
Nemunaitis J, Rao DD, Liu SH, Brunicardi FC. Nemunaitis J, et al. World J Surg. 2011 Aug;35(8):1700-14. doi: 10.1007/s00268-011-1100-0. World J Surg. 2011. PMID: 21557010 Review.
References
- Levine A. J. (1997) Cell 88, 323-331. - PubMed
- Prives C. & Hall, P. A. (1999) J. Pathol. 187, 112-126. - PubMed
- Vogelstein B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. - PubMed
- Bargonetti J. & Manfredi, J. J. (2002) Curr. Opin. Oncol. 14, 86-91. - PubMed
- Vousden K. H. (2002) Biochim. Biophys. Acta 1602, 47-59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous