Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence - PubMed (original) (raw)
Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence
Alberto Inga et al. Mol Cell Biol. 2002 Dec.
Free PMC article
Abstract
Little is known about the mechanisms that regulate differential transactivation by p53. We developed a system in the yeast Saccharomyces cerevisiae that addresses p53 transactivation capacity from 26 different p53 response elements (REs) under conditions where all other factors, such as chromatin, are kept constant. The system relies on a tightly regulated promoter (rheostatable) that can provide for a broad range of p53 expression. The p53 transactivation capacity toward each 20- to 22-bp-long RE could be ranked by using a simple phenotypic assay. Surprisingly, there was as much as a 1,000-fold difference in transactivation. There was no correlation between the functional rank and statistical predictions of binding energy of the REs. Instead we found that the central sequence element in an RE greatly affects p53 transactivation capacity, possibly because of DNA structural properties. Our results suggest that intrinsic DNA binding affinity and p53 protein levels are important contributors to p53-induced differential transactivation. These results are also relevant to understanding the regulation by other families of transcription factors that recognize several sequence-related response elements and/or have tightly regulated expression. We found that p53 had weak activity towards half the apoptotic REs. In addition, p53 alleles associated with familial breast cancer, previously classified as wild type, showed subtle differences in transactivation capacity towards several REs.
Figures
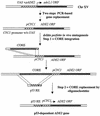
FIG. 1.
Construction of the p53-dependent ADE2 locus in the yAFM yeast strain_._ The ade2,1 open reading frame (ORF) and promoter on chromosome (Chr) XV of the yIG397 strain was first replaced with a wild-type ADE2 under control of a minimal cyc1 promoter lacking upstream activating sequences. The delitto perfetto system of in vivo mutagenesis (see text for details) was then employed to rapidly generate a panel of isogenic yeast strains whose ADE2 gene is under p53 transcriptional regulation via different p53 REs.
FIG. 2.
Regulatable expression of p53 protein by using a rheostatable GAL1 promoter. (A) pLS89 (GAL1::wild-type p53) transformants of the indicated yAFM strains were grown overnight in selective glucose medium, washed, diluted, and grown for 24 h in selective medium containing excess adenine and either raffinose or raffinose plus increasing amounts of galactose (indicated above the lanes). Different amount of protein extracts were loaded (indicated below the lanes). p53 was detected by Western blotting (with pAb1801 and DO-1). p53 induction relative to the level detected with raffinose was determined by densitometric analysis, taking into account the different amounts of protein loaded and averaging the results of the two different measurements in the case of the 0.03 and 0.12% galactose cultures. The p53 induction relative to raffinose cultures is shown for each strain. The variation in the amount of p53 among the various strains after growth on raffinose was equal to or less than threefold. (B) Relative p53 expression as a function of galactose concentration for five isogenic yAFM-RE strains. The standard deviations, the linear curve fit up to 0.12% galactose, and the correlation coefficient are shown.
FIG. 3.
Variable expression of human wild-type p53 and transactivation capacities at different p53 REs. Purified isogenic yAFM transformants with the GAL1::wild-type p53 expression vector were streaked out on plates containing raffinose as a carbon source plus increasing amount of galactose to achieve variable p53 expression. A low level of adenine in the medium was used in order to assess p53-dependent transactivation of the ADE2 gene. Colonies turned from red to pink and to white at different amounts of galactose, indicating variable activity towards the different p53 REs.
FIG. 4.
Functional ranks of 26 p53 REs. The results of the phenotypic transactivation assay at variable p53 protein expression levels are summarized. The pattern of change in colony color with increasing p53 expression is shown for every RE. Black bars, red colonies; gray bars, pink colonies; hatched bars, light pink colonies; white bars, white colonies. The y axis on the left indicates the p53 protein induction relative to the level measured on glucose. The y axis on the right indicates the galactose concentrations used in the plate assay.
FIG. 5.
Quantitative assessment of ADE2 transcription. (A) The p53 REs do not differentially affect ADE2 expression in the absence of p53. The ratio of ADE2 to ACT1 (actin) mRNA levels was determined for yAFM strains lacking the p53 expression vector as well as for the yIG397 strain, which has a wild-type ADE2 promoter. Cells were grown for 24 h in raffinose medium, followed by RNA extraction and cDNA synthesis. mRNA measurements were obtained by quantitative PCR. Plots of the real-time fluorescence measurements and a bar graph showing the relative ADE2/ACT1 mRNA ratios are shown. (B to D) The extent of ADE2 transcription depends on the p53 REs at both low and high p53 expression levels. ADE2/ACT1 mRNA ratios were determined by quantitative PCR for yAFM transformants with wild-type p53 after 24 h of growth with 2% raffinose (low p53 expression) (B), 2% galactose (C), or various galactose concentrations (expressed as a function of the relative p53 amount). The ADE2 mRNA induction is relative to the lower value detected in each experiment (RGC [B], BAX-A [C], or the level on raffinose for each strain [D]). Error bars represent the standard deviations of triplicate measurements (i.e., three independent cultures and RNA preparations). A Western blot showing the variation of p53 expression on 2% galactose is also shown in panel C. For each strain, 5 μg of extract from raffinose cultures and 1 and 5 μg from galactose cultures were loaded.
FIG. 5.
Quantitative assessment of ADE2 transcription. (A) The p53 REs do not differentially affect ADE2 expression in the absence of p53. The ratio of ADE2 to ACT1 (actin) mRNA levels was determined for yAFM strains lacking the p53 expression vector as well as for the yIG397 strain, which has a wild-type ADE2 promoter. Cells were grown for 24 h in raffinose medium, followed by RNA extraction and cDNA synthesis. mRNA measurements were obtained by quantitative PCR. Plots of the real-time fluorescence measurements and a bar graph showing the relative ADE2/ACT1 mRNA ratios are shown. (B to D) The extent of ADE2 transcription depends on the p53 REs at both low and high p53 expression levels. ADE2/ACT1 mRNA ratios were determined by quantitative PCR for yAFM transformants with wild-type p53 after 24 h of growth with 2% raffinose (low p53 expression) (B), 2% galactose (C), or various galactose concentrations (expressed as a function of the relative p53 amount). The ADE2 mRNA induction is relative to the lower value detected in each experiment (RGC [B], BAX-A [C], or the level on raffinose for each strain [D]). Error bars represent the standard deviations of triplicate measurements (i.e., three independent cultures and RNA preparations). A Western blot showing the variation of p53 expression on 2% galactose is also shown in panel C. For each strain, 5 μg of extract from raffinose cultures and 1 and 5 μg from galactose cultures were loaded.
FIG. 5.
Quantitative assessment of ADE2 transcription. (A) The p53 REs do not differentially affect ADE2 expression in the absence of p53. The ratio of ADE2 to ACT1 (actin) mRNA levels was determined for yAFM strains lacking the p53 expression vector as well as for the yIG397 strain, which has a wild-type ADE2 promoter. Cells were grown for 24 h in raffinose medium, followed by RNA extraction and cDNA synthesis. mRNA measurements were obtained by quantitative PCR. Plots of the real-time fluorescence measurements and a bar graph showing the relative ADE2/ACT1 mRNA ratios are shown. (B to D) The extent of ADE2 transcription depends on the p53 REs at both low and high p53 expression levels. ADE2/ACT1 mRNA ratios were determined by quantitative PCR for yAFM transformants with wild-type p53 after 24 h of growth with 2% raffinose (low p53 expression) (B), 2% galactose (C), or various galactose concentrations (expressed as a function of the relative p53 amount). The ADE2 mRNA induction is relative to the lower value detected in each experiment (RGC [B], BAX-A [C], or the level on raffinose for each strain [D]). Error bars represent the standard deviations of triplicate measurements (i.e., three independent cultures and RNA preparations). A Western blot showing the variation of p53 expression on 2% galactose is also shown in panel C. For each strain, 5 μg of extract from raffinose cultures and 1 and 5 μg from galactose cultures were loaded.
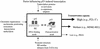
FIG. 6.
Intrinsic DNA binding and differential transactivation by p53. Many factors can influence p53 transactivation (arrows). In this study, the factors are kept constant by using isogenic yeast in the functional assay. Thus, a difference in transactivation capacity solely reflects differences in the intrinsic DNA binding affinity of p53 for the individual REs (box). Following sequence-specific DNA binding, p53 may undergo conformational changes that alter tetramer stability and favor protein-protein interaction with the basal transcriptional machinery.
Similar articles
- Noncanonical DNA motifs as transactivation targets by wild type and mutant p53.
Jordan JJ, Menendez D, Inga A, Noureddine M, Bell DA, Resnick MA. Jordan JJ, et al. PLoS Genet. 2008 Jun 27;4(6):e1000104. doi: 10.1371/journal.pgen.1000104. PLoS Genet. 2008. PMID: 18714371 Free PMC article. - Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation.
Jordan JJ, Inga A, Conway K, Edmiston S, Carey LA, Wu L, Resnick MA. Jordan JJ, et al. Mol Cancer Res. 2010 May;8(5):701-16. doi: 10.1158/1541-7786.MCR-09-0442. Epub 2010 Apr 20. Mol Cancer Res. 2010. PMID: 20407015 Free PMC article. Clinical Trial. - Potentiating the p53 network.
Menendez D, Inga A, Resnick MA. Menendez D, et al. Discov Med. 2010 Jul;10(50):94-100. Discov Med. 2010. PMID: 20670604 Review. - The expanding universe of p53 targets.
Menendez D, Inga A, Resnick MA. Menendez D, et al. Nat Rev Cancer. 2009 Oct;9(10):724-37. doi: 10.1038/nrc2730. Nat Rev Cancer. 2009. PMID: 19776742 Review.
Cited by
- Delineation of the DNA Structural Features of Eukaryotic Core Promoter Classes.
Vanaja A, Yella VR. Vanaja A, et al. ACS Omega. 2022 Feb 9;7(7):5657-5669. doi: 10.1021/acsomega.1c04603. eCollection 2022 Feb 22. ACS Omega. 2022. PMID: 35224327 Free PMC article. - Inhibition of p53 and/or AKT as a new therapeutic approach specifically targeting ALT cancers.
Ge Y, Wu S, Zhang Z, Li X, Li F, Yan S, Liu H, Huang J, Zhao Y. Ge Y, et al. Protein Cell. 2019 Nov;10(11):808-824. doi: 10.1007/s13238-019-0634-z. Epub 2019 May 21. Protein Cell. 2019. PMID: 31115790 Free PMC article. - NEAT1 Long Isoform Is Highly Expressed in Chronic Lymphocytic Leukemia Irrespectively of Cytogenetic Groups or Clinical Outcome.
Ronchetti D, Favasuli V, Monti P, Cutrona G, Fabris S, Silvestris I, Agnelli L, Colombo M, Menichini P, Matis S, Gentile M, Nurtdinov R, Guigó R, Baldini L, Fronza G, Ferrarini M, Morabito F, Neri A, Taiana E. Ronchetti D, et al. Noncoding RNA. 2020 Mar 13;6(1):11. doi: 10.3390/ncrna6010011. Noncoding RNA. 2020. PMID: 32182990 Free PMC article. - Differential gene expression in primary human skin keratinocytes and fibroblasts in response to ionizing radiation.
Warters RL, Packard AT, Kramer GF, Gaffney DK, Moos PJ. Warters RL, et al. Radiat Res. 2009 Jul;172(1):82-95. doi: 10.1667/RR1677.1. Radiat Res. 2009. PMID: 19580510 Free PMC article. - Dose-dependent dual role of PIT-1 (POU1F1) in somatolactotroph cell proliferation and apoptosis.
Jullien N, Roche C, Brue T, Figarella-Branger D, Graillon T, Barlier A, Herman JP. Jullien N, et al. PLoS One. 2015 Mar 30;10(3):e0120010. doi: 10.1371/journal.pone.0120010. eCollection 2015. PLoS One. 2015. PMID: 25822178 Free PMC article.
References
- Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses Eur. J. Biochem. 268:2764-2772. - PubMed
- Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. - PubMed
- Berg, O. G., and P. H. von Hippel. 1987. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 193:723-750. - PubMed
- Biggin, M. D. 2001. To bind or not to bind. Nat. Genet. 28:303-304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous