Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase - PubMed (original) (raw)
Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase
Yuichiro Shimizu et al. EMBO J. 2003.
Abstract
The XPC-HR23B complex recognizes various helix-distorting lesions in DNA and initiates global genome nucleotide excision repair. Here we describe a novel functional interaction between XPC-HR23B and thymine DNA glycosylase (TDG), which initiates base excision repair (BER) of G/T mismatches generated by spontaneous deamination of 5-methylcytosine. XPC-HR23B stimulated TDG activity by promoting the release of TDG from abasic sites that result from the excision of mismatched T bases. In the presence of AP endonuclease (APE), XPC-HR23B had an additive effect on the enzymatic turnover of TDG without significantly inhibiting the subsequent action of APE. Our observations suggest that XPC-HR23B may participate in BER of G/T mismatches, thereby contributing to the suppression of spontaneous mutations that may be one of the contributory factors for the promotion of carcinogenesis in xeroderma pigmentosum genetic complementation group C patients.
Figures
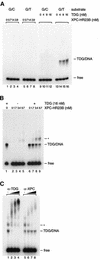
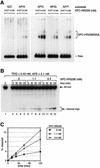
Fig. 1. Physical interaction of XPC–HR23B and TDG. Glutathione–Sepharose beads (20 µl) were incubated in 100 µl of the binding mixture with 16.6 nM of either GST (lane 3) or GST–TDG (lanes 4–6) in the presence of XPC–HR23B (6.7 nM). For lanes 5 and 6, 166 nM and 1.66 µM His-TDG, respectively, were also included as a competitor. After extensive washing, bound proteins were eluted with buffer containing 10 mM glutathione. A quarter of each eluate was mixed with whole-cell extract from XP4PASV cells, which do not express XPC, and subjected to 8% SDS–PAGE followed by immunoblotting with anti-XPC antibody. Lane 1, 0.4% of the input XPC; lane 2, XP4PASV cell extract only.
Fig. 2. DNA substrates used in gel mobility shift analysis (A) and the nicking assays (B). X is A, C, G or T.
Fig. 3. XPC–HR23B is recruited to TDG-bound DNA and forms a ternary complex. (A) The indicated concentrations of XPC–HR23B or TDG were incubated at 30°C for 30 min with 0.35 nM of the 32P-labeled DNA fragment containing a single G/T mismatch or completely paired double-stranded DNA (G/C) as a control. The resulting DNA–protein complex was fixed with glutaraldehyde, and resolved by native PAGE. (B) The DNA substrate containing a single G/T mismatch (0.35 nM) was incubated at 30°C for 15 min in the presence or absence of His-TDG (16 nM). The indicated concentrations of XPC–HR23B were then added, incubated further at 30°C for 15 min and subjected to native PAGE after glutaraldehyde fixation. The asterisk shows the bands that newly appeared in the presence of both TDG and XPC–HR23B. (C) A supershift assay identifying the ternary complex containing TDG, XPC–HR23B and DNA. Sequential binding reactions were conducted as in (B) with 16 nM TDG, 6.7 nM XPC–HR23B and 0.35 nM DNA substrate containing a single G/T mismatch. Various amounts of anti-TDG or anti-XPC antibodies were then added and incubated further on ice for 15 min before cross-linking the DNA–protein complexes with glutaraldehyde.
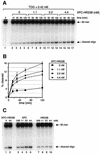
Fig. 4. XPC–HR23B stimulates the activity of TDG. (A) A nicking assay measuring the TDG activity in the presence of XPC–HR23B. The 60mer DNA substrate containing a single G/T mismatch, which was labeled at the 5′ end of the T-strand, was incubated at 30°C for the indicated time with 0.42 nM His-TDG in the presence or absence of various concentrations of XPC–HR23B. The DNA samples were purified and subjected to alkali treatment to cleave the resulting AP sites, after which denaturing PAGE was performed. (B) The percentage of the cleaved oligonucleotides in the 32P-labeled DNA substrate was calculated for each lane in (A) and plotted as a graph. The mean values and standard errors were calculated from at least two independent experiments. (C) A nicking assay using 0.42 nM His-TDG in the presence of various concentrations of XPC–HR23B (lane 2), XPC alone (lanes 3–6) or HR23B alone (lanes 7–10) as indicated. All reactions were incubated at 30°C for 120 min and the purified DNA samples were subjected to alkali treatment and denaturing PAGE.
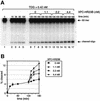
Fig. 5. XPC–HR23B promotes the turnover of TDG. (A) The 32P-labeled DNA substrate containing a single G/T mismatch was incubated with 0.42 nM His-TDG at 30°C for various periods as indicated (lanes 1–5). After 120 min, the indicated concentrations of XPC–HR23B were added and incubated further at 30°C for 15, 30 or 60 min (total incubation time is shown above each lane). The purified DNA samples were subjected to alkali treatment and denaturing PAGE. (B) The percentage of the cleaved oligonucleotides in the 32P-labeled DNA substrate was calculated for each lane in (A). The mean values and standard errors were calculated from at least two independent experiments.
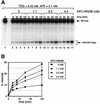
Fig. 6. XPC–HR23B stimulates the activity of TDG in the presence of APE. (A) The 32P-labeled DNA substrate containing a single G/T mismatch was incubated with 0.42 nM His-TDG, 2.1 nM His-APE and the indicated concentrations of XPC–HR23B at 30°C for the specified time. The purified DNA samples were subjected to alkali treatment and denaturing PAGE. Due to the 3′→5′ exonuclease activity of APE, the bands of cleaved oligonucleotides appear as ladders. (B) The percentage of the cleaved oligonucleotides in the 32P-labeled DNA substrate was calculated for each lane in (A). The mean values and standard errors were calculated from at least two independent experiments.
Fig. 7. XPC–HR23B does not inhibit the enzymatic action of APE. (A) The 32P-labeled DNA substrate containing a synthetic AP site analog opposite four different bases or a normal G/C pair at the corresponding position (0.35 nM each) were incubated at 30°C for 30 min with the indicated concentrations of XPC–HR23B. The resulting DNA–protein complex was fixed with glutaraldehyde, and resolved by native PAGE. (B) The 32P-labeled DNA substrate containing a single G/T mismatch was incubated with 0.42 nM His-TDG, 2.1 nM His-APE and the indicated concentrations of XPC–HR23B at 30°C for the specified periods. The reactions were stopped by addition of EDTA and subjected to denaturing PAGE without alkali treatment. (C) The percentage of the cleaved oligonucleotides in the 32P-labeled DNA substrate was calculated for each lane in (B). The mean values and standard errors were calculated from at least two independent experiments.
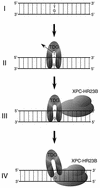
Fig. 8. A schematic model depicting a possible mechanism for TDG stimulation by XPC–HR23B. A G/T mismatch can be generated by spontaneous deamination of 5-methylcytosine (I). After the removal of the mismatched T (II), TDG remains attached to the resulting AP site through interaction with the opposite guanine residue. XPC–HR23B is then recruited via protein–protein interaction and/or structural changes of DNA induced by TDG (III). It has been shown that XPC–HR23B preferentially recognizes DNA secondary structure containing a junction with a single-stranded arm, branching away from duplex DNA in the 3′ direction (Sugasawa et al., 2002). Therefore, XPC–HR23B probably interacts with the unpaired guanine and binds to the 3′ side of the AP site. XPC–HR23B may thus compete with TDG for the guanine residue and push it out from the AP site (IV).
Similar articles
- Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover.
Hardeland U, Steinacher R, Jiricny J, Schär P. Hardeland U, et al. EMBO J. 2002 Mar 15;21(6):1456-64. doi: 10.1093/emboj/21.6.1456. EMBO J. 2002. PMID: 11889051 Free PMC article. - Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases.
Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Mol CD, et al. J Mol Biol. 2002 Jan 18;315(3):373-84. doi: 10.1006/jmbi.2001.5264. J Mol Biol. 2002. PMID: 11786018 - Nucleotide excision repair of 5-formyluracil in vitro is enhanced by the presence of mismatched bases.
Kino K, Shimizu Y, Sugasawa K, Sugiyama H, Hanaoka F. Kino K, et al. Biochemistry. 2004 Mar 16;43(10):2682-7. doi: 10.1021/bi0361416. Biochemistry. 2004. PMID: 15005603 - Thymine DNA glycosylase.
Hardeland U, Bentele M, Lettieri T, Steinacher R, Jiricny J, Schär P. Hardeland U, et al. Prog Nucleic Acid Res Mol Biol. 2001;68:235-53. doi: 10.1016/s0079-6603(01)68103-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554300 Review. - Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein.
Clement FC, Camenisch U, Fei J, Kaczmarek N, Mathieu N, Naegeli H. Clement FC, et al. Mutat Res. 2010 Mar 1;685(1-2):21-8. doi: 10.1016/j.mrfmmm.2009.08.005. Epub 2009 Aug 15. Mutat Res. 2010. PMID: 19686765 Review.
Cited by
- Xeroderma Pigmentosum Complementation Group C (XPC): Emerging Roles in Non-Dermatologic Malignancies.
Nasrallah NA, Wiese BM, Sears CR. Nasrallah NA, et al. Front Oncol. 2022 Apr 21;12:846965. doi: 10.3389/fonc.2022.846965. eCollection 2022. Front Oncol. 2022. PMID: 35530314 Free PMC article. Review. - Kinetic Analysis of the Effect of _N_-Terminal Acetylation on Thymine DNA Glycosylase.
Tarantino ME, Delaney S. Tarantino ME, et al. Biochemistry. 2022 May 17;61(10):895-908. doi: 10.1021/acs.biochem.1c00823. Epub 2022 Apr 18. Biochemistry. 2022. PMID: 35436101 Free PMC article. - The correlation between polymorphisms in the XPC gene and glioma susceptibility in a Chinese pediatric population.
Zhang Z, Huang Y, Chen H, Wu P, Deng Z, Deng G, Zheng Y, Li G, Yuan L, Xu Y. Zhang Z, et al. Transl Pediatr. 2021 Jul;10(7):1896-1904. doi: 10.21037/tp-21-301. Transl Pediatr. 2021. PMID: 34430438 Free PMC article. - Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.
Paul D, Mu H, Tavakoli A, Dai Q, Chakraborty S, He C, Ansari A, Broyde S, Min JH. Paul D, et al. DNA Repair (Amst). 2021 Nov;107:103194. doi: 10.1016/j.dnarep.2021.103194. Epub 2021 Jul 29. DNA Repair (Amst). 2021. PMID: 34428697 Free PMC article. - Every protagonist has a sidekick: Structural aspects of human xeroderma pigmentosum-binding proteins in nucleotide excision repair.
Feltes BC. Feltes BC. Protein Sci. 2021 Nov;30(11):2187-2205. doi: 10.1002/pro.4173. Epub 2021 Aug 27. Protein Sci. 2021. PMID: 34420242 Free PMC article. Review.
References
- Araki M., Masutani,C., Takemura,M., Uchida,A., Sugasawa,K., Kondoh,J., Ohkuma,Y. and Hanaoka,F. (2001) Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem., 276, 18665–18672. - PubMed
- Araújo S.J., Tirode,F., Coin,F., Pospiech,H., Syväoja,J.E., Stucki,M., Hübscher,U., Egly,J.-M. and Wood,R.D. (2000) Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH and modulation by CAK. Genes Dev., 14, 349–359. - PMC - PubMed
- Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous