Analysis of helicase activity and substrate specificity of Drosophila RECQ5 - PubMed (original) (raw)
Analysis of helicase activity and substrate specificity of Drosophila RECQ5
A Zeynep Ozsoy et al. Nucleic Acids Res. 2003.
Abstract
RecQ5 is one of five RecQ helicase homologs identified in humans. Three of the human RecQ homologs (BLM, WRN and RTS) have been linked to autosomal recessive human genetic disorders (Bloom syndrome, Werner syndrome and Rothmund-Thomson syndrome, respectively) that display increased genomic instability and cause elevated levels of cancers in addition to other symptoms. To understand the role of RecQ helicases in maintaining genomic stability, the WRN, BLM and Escherichia coli RecQ helicases have been characterized in terms of their DNA substrate specificity. However, little is known about other members of the RecQ family. Here we show that Drosophila RECQ5 helicase is a structure-specific DNA helicase like the other RecQ helicases biochemically characterized so far, although the substrate specificity is not identical to that of WRN and BLM helicases. Drosophila RECQ5 helicase is capable of unwinding 3' Flap, three-way junction, fork and three-strand junction substrates at lower protein concentrations compared to 5' Flap, 12 nt bubble and synthetic Holliday junction structures, which can be unwound efficiently by WRN and BLM.
Figures
Figure 1
Unwinding of DNA substrates by DmRECQ5. Helicase assays using the 3′ Flap (open circles), 5′ Flap (open triangles), three-way junction (filled squares), 50/19 partial duplex (filled circles) and fork (fork 1, open squares; fork 2, filled triangles) substrates were as described in Materials and Methods using the indicated amount of DmRECQ5. Unwinding reactions were initiated by the addition of enzyme and incubated for 10 min at 30°C. Each data point is the average of at least three experiments. Error bars represent the standard deviation about the mean.
Figure 2
Unwinding of a synthetic Holliday junction substrate. Helicase assays using a synthetic Holliday junction substrate were as described in Materials and Methods using the indicated amount of DmRECQ5 and MgCl2. Unwinding reactions were initiated by the addition of enzyme and incubated for 10 min at 30°C. Each data point is the average of at least two experiments.
Figure 3
Unwinding of the 3′ Flap substrate. (A) A DmRECQ5 titration using the 3′ Flap substrate. Lanes 1 and 2 contained no enzyme. The reaction shown in lane 2 was boiled to denature the substrate. Lane 3, 910 nM DmRECQ5; lane 4, 455 nM DmRECQ5; lane 5, 226 nM DmRECQ5; lane 6, 113 nM DmRECQ5, lane 7, 57 nM DmRECQ5; lane 8, 28 nM DmRECQ5; lane 9, 14 nM DmRECQ5; lane 10, 7 nM DmRECQ5; lane 11, 3.6 nM DmRECQ5; lane 12, 1.8 nM DmRECQ5. Lanes 13 and 14 are markers to indicate where the fork and the partial duplex structures migrate on the gel, respectively. The substrate and each intermediate/product are shown schematically on the right. (B) A schematic representation of unwinding of the 3′ Flap substrate. Production of two intermediates can be explained by two pathways. The partial duplex intermediate is produced by the upper pathway when the protein loads on the ssDNA 3′ flap and translocates along this strand displacing the top strand from the bottom strand. The fork intermediate is produced by the lower pathway when the protein loads on the bottom strand at the junction translocating 3′ to 5′ along this strand unwinding the annealed oligonucleotide. (C) Time course of 3′ Flap unwinding. The reaction mixture (100 µl) was as described in Materials and Methods and the reaction was initiated by the addition of RECQ5 to a final concentration of 53 nM. Aliquots (10 µl) of the reaction were withdrawn at the indicated time points, quenched and analyzed on a native polyacrylamide gel. Lines correspond to fraction of fork (open circles), partial duplex (filled triangles) and 50mer (filled circles) present during the time course. The arrow points to the 20 s time point when the appearance of the final product (50mer) is first observed.
Figure 4
Unwinding of the three-way junction substrate. (A) A DmRECQ5 titration using the three-way junction substrate. Lanes 1 and 2 contained no enzyme. The reaction shown in lane 2 was boiled prior to loading on the gel to denature the substrate. Lanes 3–8 show a protein titration (1.8, 3.5, 7, 14, 28, 57 nM, respectively). Lanes 9–12 are markers to indicate where the 3′ Flap, 5′ Flap, Fork1 and the 50/19 partial duplex structures migrate on the gel, repectively. (B) A schematic representation of unwinding of the three-way junction substrate. Production of the 5′ Flap structure from the three-way junction substrate can be achieved if RECQ5 loads on the bottom strand at the junction and translocates along this strand displacing the annealed oligonucleotide. At high protein concentrations some of the 5′ Flap intermediate is unwound to release the labeled 50mer. This activity is shown by a thin arrow compared to the thicker arrow to produce the 5′ Flap intermediate from the three-way junction substrate.
Figure 5
EMSA of the three-way junction substrate. DNA binding reactions were as described in Materials and Methods. Lanes 1–7, reactions contained the three-way junction substrate; lanes 8–14, reactions contained the 80 bp blunt duplex substrate. Lanes 1 and 8 contained no DmRECQ5; lanes 2 and 9 contained 7.3 nM DmRECQ5; lanes 3 and 10 contained 14.5 nM DmRECQ5; lanes 4 and 11 contained 29 nM DmRECQ5; lanes 5 and 12 contained 58 nM DmRECQ5; lanes 6 and 13 contained 116 nM DmRECQ5; lanes 7 and 14 contained 232 nM DmRECQ5. Schematics of each substrate are shown at the top of the corresponding gel.
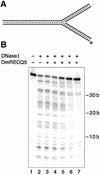
Figure 6
DNase I footprinting on the three-way junction substrate. DNase I footprinting reaction mixtures were prepared and analyzed as described in Materials and Methods. (A) Schematic view of the DNA substrate. The labeled strand is marked with the asterisk. (B) The reaction in lane 1 contained no enzymes, the reaction in lane 2 contained no DmRECQ5 and lanes 3–7 contained DmRECQ5 at 3.8, 7.3, 14.6, 29.1 and 58.3 nM, respectively.
Figure 7
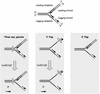
The three-way junction, 3′ Flap and 5′ Flap substrates resemble a stalled replication fork. A replication fork is illustrated at the top to describe all the strands involved and their polarities. The three panels illustrate the observed unwinding pathway for each substrate. The hatched box on the three-way junction and the 3′ Flap structures denotes a lesion that blocks replication. The solid lines represent the oligonucleotides present in each substrate used. Dashed lines are drawn to further illustrate the proposed replication fork. The three-way junction mimics a fork where a lesion blocks synthesis on both sides. The 3′ Flap mimics the fork where DNA synthesis of the leading strand is blocked. The 5′ Flap mimics the fork where DNA synthesis of the lagging strand has been blocked or has not yet reached the point of the leading strand, due to uncoupling of the leading and lagging strand synthesis. The resulting structures from the RECQ5-catalyzed unwinding reaction are depicted on the bottom. DmRECQ5 removes the lagging strand in the three-way junction and 3′ Flap substrates.
Similar articles
- Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins.
Machwe A, Lozada EM, Xiao L, Orren DK. Machwe A, et al. BMC Mol Biol. 2006 Jan 13;7:1. doi: 10.1186/1471-2199-7-1. BMC Mol Biol. 2006. PMID: 16412221 Free PMC article. - Enzymatic mechanism of the WRN helicase/nuclease.
Brosh RM Jr, Opresko PL, Bohr VA. Brosh RM Jr, et al. Methods Enzymol. 2006;409:52-85. doi: 10.1016/S0076-6879(05)09004-X. Methods Enzymol. 2006. PMID: 16793395 - The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases.
Mohaghegh P, Karow JK, Brosh RM Jr, Bohr VA, Hickson ID. Mohaghegh P, et al. Nucleic Acids Res. 2001 Jul 1;29(13):2843-9. doi: 10.1093/nar/29.13.2843. Nucleic Acids Res. 2001. PMID: 11433031 Free PMC article. - Human RecQL4 helicase plays multifaceted roles in the genomic stability of normal and cancer cells.
Mo D, Zhao Y, Balajee AS. Mo D, et al. Cancer Lett. 2018 Jan 28;413:1-10. doi: 10.1016/j.canlet.2017.10.021. Epub 2017 Nov 7. Cancer Lett. 2018. PMID: 29080750 Review. - RecQ family helicases: roles as tumor suppressor proteins.
Nakayama H. Nakayama H. Oncogene. 2002 Dec 16;21(58):9008-21. doi: 10.1038/sj.onc.1205959. Oncogene. 2002. PMID: 12483516 Review.
Cited by
- RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures.
Popuri V, Croteau DL, Brosh RM Jr, Bohr VA. Popuri V, et al. Cell Cycle. 2012 Nov 15;11(22):4252-65. doi: 10.4161/cc.22581. Epub 2012 Oct 24. Cell Cycle. 2012. PMID: 23095637 Free PMC article. - Roles of Werner syndrome protein in protection of genome integrity.
Rossi ML, Ghosh AK, Bohr VA. Rossi ML, et al. DNA Repair (Amst). 2010 Mar 2;9(3):331-44. doi: 10.1016/j.dnarep.2009.12.011. Epub 2010 Jan 13. DNA Repair (Amst). 2010. PMID: 20075015 Free PMC article. Review. - RecQ helicases: multifunctional genome caretakers.
Chu WK, Hickson ID. Chu WK, et al. Nat Rev Cancer. 2009 Sep;9(9):644-54. doi: 10.1038/nrc2682. Epub 2009 Aug 6. Nat Rev Cancer. 2009. PMID: 19657341 Review. - Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery.
Tuteja N, Tuteja R. Tuteja N, et al. Eur J Biochem. 2004 May;271(10):1835-48. doi: 10.1111/j.1432-1033.2004.04093.x. Eur J Biochem. 2004. PMID: 15128294 Free PMC article. Review. - Biochemical characterization of Warsaw breakage syndrome helicase.
Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM Jr. Wu Y, et al. J Biol Chem. 2012 Jan 6;287(2):1007-21. doi: 10.1074/jbc.M111.276022. Epub 2011 Nov 18. J Biol Chem. 2012. PMID: 22102414 Free PMC article.
References
- Ellis N.A., Groden,J., Ye,T.-Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. - PubMed
- Yu C.-E., Oshima,J., Fu,Y.-H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. - PubMed
- Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson Syndrome. Nature Genet., 22, 82–84. - PubMed
- van Brabant A.J., Stan,R. and Ellis,N.A. (2000) DNA helicases, genomic instability and human genetic disease. Annu. Rev. Genom. Hum. Genet., 1, 409–459. - PubMed
- Puranam K.L. and Blackshear,P.J. (1994) Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem., 269, 29838–29845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials