Lentivirus-delivered stable gene silencing by RNAi in primary cells - PubMed (original) (raw)
doi: 10.1261/rna.2192803.
Derek M Dykxhoorn, Deborah Palliser, Hana Mizuno, Evan Y Yu, Dong Sung An, David M Sabatini, Irvin S Y Chen, William C Hahn, Phillip A Sharp, Robert A Weinberg, Carl D Novina
Affiliations
- PMID: 12649500
- PMCID: PMC1370415
- DOI: 10.1261/rna.2192803
Lentivirus-delivered stable gene silencing by RNAi in primary cells
Sheila A Stewart et al. RNA. 2003 Apr.
Abstract
Genome-wide genetic approaches have proven useful for examining pathways of biological significance in model organisms such as Saccharomyces cerevisiae, Drosophila melanogastor, and Caenorhabditis elegans, but similar techniques have proven difficult to apply to mammalian systems. Although manipulation of the murine genome has led to identification of genes and their function, this approach is laborious, expensive, and often leads to lethal phenotypes. RNA interference (RNAi) is an evolutionarily conserved process of gene silencing that has become a powerful tool for investigating gene function by reverse genetics. Here we describe the delivery of cassettes expressing hairpin RNA targeting green fluorescent protein (GFP) using Moloney leukemia virus-based and lentivirus-based retroviral vectors. Both transformed cell lines and primary dendritic cells, normally refractory to transfection-based gene transfer, demonstrated stable silencing of targeted genes, including the tumor suppressor gene TP53 in normal human fibroblasts. This report demonstrates that both Moloney leukemia virus and lentivirus vector-mediated expression of RNAi can achieve effective, stable gene silencing in diverse biological systems and will assist in elucidating gene functions in numerous cell types including primary cells.
Figures
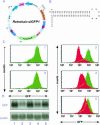
FIGURE 1.
Sequence-specific knockdown of GFP expression by retrovirus-delivered GFP hairpin RNA expression. (A) Schematic presentation of Retrohair-slGFP, a replication incompetent retrovirus that contains a pol-III promoter (U6), a stem-loop cassette followed by a 5T pol-III termination signal, cloned 3′ to a truncated gag lacking a translation start sequence (ATG). Retrohair can be selected by addition of puromycin. (B) The stem-loop encoded hairpin RNA has sequence identity to a 21-nt region of GFP mRNA. (C) FACs analysis of GFP expression in HeLa-GFP infected with (1 and 3) empty vector controls, (2) Retrohair-slGFP1, (4) Retrohair-slGFP2, or (5) Retrohair-slGFP2mut. Overlay histograms in panels 2, 4, and 5 demonstrate the level of knockdown in cells infected with Retrohair-containing stem-loop cassettes (red) compared to cells infected with empty vector controls (green). (D) Northern blot analysis of GFP expression in HeLa-GFP cells described in C using a GFP-specific probe. Northern blot with a β-actin probe served as a loading control.
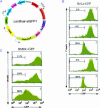
FIGURE 2.
Time course of selection of GFP knockdown in RNAi-stable HeLa-GFP cells. (A) FACS analysis of GFP expression in HeLa-GFP cells infected with Retrohair-lacking a stem-loop (empty, top panels) or with Retrohair-slGFP1 (-slGFP1, bottom panels). GFP expression was measured in HeLa-GFP either preselection, with 3 d, 2 wk, or 4 wk of puromycin selection or in HeLa-GFP after 2 wk of growth with puromycin selection followed by 2 wk of growth without puromycin selection. (B) Northern blot analysis of GFP expression in HeLa (lane C), in HeLa-GFP infected with Retrohair at (lane 1) 3 d (lane 2) 2 wk, (lane 3) 4 wk, or infected with Retrohair-slGFP1 at (lane 4) 3 d (lane 5) 2 wk, (lane 6) 4 wk, or at (lane 7) 2 wk with selection and 2 wk without puromycin selection. β-Actin expression served as a loading control. (C) Modified Northern blot analysis of the samples described in B using an RNA probe (sense strand) with complementarity to the antisense strand of the GFP hairpin RNA. Samples were normalized by total RNA content. Titration of GFP siRNA was used as a size marker and for quantitation by densitometry of small RNAs. (D) FACS analysis of GFP expression in HeLa-GFP infected with Retrohair-slGFP1 4 wk after selection on puromycin (green) or infected with Retrohair-slGFP1 4 wk after selection on puromycin and then superinfected with Retrohair-slGFP2 (red).
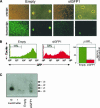
FIGURE 3.
Knockdown of GFP expression in DC-GFP. (A) Photomicroscopy of DC-GFP visualized by white light or by fluorescent light after infection with Retrohair or with Retrohair-slGFP1(slGFP1). (B) FACS of GFP expression in DC-GFP infected with the Retrohair or Retrohair-slGFP1 retrovirus. A bar graph depicts the percent decrease in GFP expression as measured by MFI. Error bars represent the average (±SD) of six experiments. (C) Modified Northern blot analysis of Retrohair and Retrohair-slGFP1-infected DC-GFP using an RNA probe (sense strand) with complementarity to the antisense strand of the GFP hairpin RNA. Samples were normalized by total RNA content. Titration of GFP siRNA was used as size marker and for quantitation by densitometry of small RNAs.
FIGURE 4.
Knockdown of GFP expression by lentivirus-delivered RNAi. (A) Schematic presentation of the lentiviral construct. Lentihair is a replication incompetent, self-inactivating retrovirus generated by cloning the U6 promoter and stem-loop cassette targeted to luciferase (Lentihair-slLUC) or GFP (Lentihair-slGFP1) 5′ of the cPPT. A puromycin resistance gene was cloned 3′ of the human hPGK promoter. (B) FACS analysis of GFP expression in HeLa-GFP cells (1) uninfected or infected with (2) Retrohair, (3) Retrohair-slGFP1, (4) Lentihair-slLUC or (5) Lentihair-slGFP. (C) FACS analysis of GFP expression in (1) uninfected, (2) Lentihair-slLUC, or (3) Lentihair-slGFP infected DC-GFP.
Similar articles
- Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi.
Sapru MK, Yates JW, Hogan S, Jiang L, Halter J, Bohn MC. Sapru MK, et al. Exp Neurol. 2006 Apr;198(2):382-90. doi: 10.1016/j.expneurol.2005.12.024. Epub 2006 Feb 7. Exp Neurol. 2006. PMID: 16455076 - First knockdown gene expression in bat (Hipposideros armiger) brain mediated by lentivirus.
Chen Q, Zhu T, Jones G, Zhang J, Sun Y. Chen Q, et al. Mol Biotechnol. 2013 Jun;54(2):564-71. doi: 10.1007/s12033-012-9596-6. Mol Biotechnol. 2013. PMID: 22965420 - Knockdown of human p53 gene expression in 293-T cells by retroviral vector-mediated short hairpin RNA.
Hao DL, Liu CM, Dong WJ, Gong H, Wu XS, Liu DP, Liang CC. Hao DL, et al. Acta Biochim Biophys Sin (Shanghai). 2005 Nov;37(11):779-83. doi: 10.1111/j.1745-7270.2005.00107.x. Acta Biochim Biophys Sin (Shanghai). 2005. PMID: 16270158 - Mammalian RNAi: a practical guide.
Sandy P, Ventura A, Jacks T. Sandy P, et al. Biotechniques. 2005 Aug;39(2):215-24. doi: 10.2144/05392RV01. Biotechniques. 2005. PMID: 16116795 Review. - miRNA cassettes in viral vectors: problems and solutions.
Liu YP, Berkhout B. Liu YP, et al. Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):732-45. doi: 10.1016/j.bbagrm.2011.05.014. Epub 2011 Jun 7. Biochim Biophys Acta. 2011. PMID: 21679781 Review.
Cited by
- Oncolytic viruses alter the biogenesis of tumor extracellular vesicles and influence their immunogenicity.
Hirigoyen U, Guilbaud C, Krejbich M, Fouet M, Fresquet J, Arnaud B, Com E, Pineau C, Cadiou G, Burlaud-Gaillard J, Erbs P, Fradin D, Labarrière N, Fonteneau JF, Petithomme T, Boisgerault N. Hirigoyen U, et al. Mol Ther Oncol. 2024 Sep 26;32(4):200887. doi: 10.1016/j.omton.2024.200887. eCollection 2024 Dec 19. Mol Ther Oncol. 2024. PMID: 39492948 Free PMC article. - Expanding Insights: Harnessing Expansion Microscopy for Super-Resolution Analysis of HIV-1-Cell Interactions.
Petrich A, Hwang GM, La Rocca L, Hassan M, Anders-Össwein M, Sonntag-Buck V, Heuser AM, Laketa V, Müller B, Kräusslich HG, Klaus S. Petrich A, et al. Viruses. 2024 Oct 15;16(10):1610. doi: 10.3390/v16101610. Viruses. 2024. PMID: 39459943 Free PMC article. - Investigation of Cell Mechanics and Migration on DDR2-Expressing Neuroblastoma Cell Line.
Vessella T, Rozen EJ, Shohet J, Wen Q, Zhou HS. Vessella T, et al. Life (Basel). 2024 Oct 2;14(10):1260. doi: 10.3390/life14101260. Life (Basel). 2024. PMID: 39459560 Free PMC article. - Parallel genome-scale CRISPR-Cas9 screens uncouple human pluripotent stem cell identity versus fitness.
Rosen BP, Li QV, Cho HS, Liu D, Yang D, Graff S, Yan J, Luo R, Verma N, Damodaran JR, Kale HT, Kaplan SJ, Beer MA, Sidoli S, Huangfu D. Rosen BP, et al. Nat Commun. 2024 Oct 17;15(1):8966. doi: 10.1038/s41467-024-53284-4. Nat Commun. 2024. PMID: 39419994 Free PMC article. - Epstein-Barr virus induces host shutoff extensively via BGLF5-independent mechanisms.
Casco A, Ohashi M, Johannsen E. Casco A, et al. Cell Rep. 2024 Oct 22;43(10):114743. doi: 10.1016/j.celrep.2024.114743. Epub 2024 Sep 18. Cell Rep. 2024. PMID: 39298313 Free PMC article.
References
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002a. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. - PubMed
- ———. 2002b. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. Published online. - PubMed
- Burns, J.C., Friedmann, T., Driever, W., Burrascano, M., and Yee, J.K. 1993. Vesicular stomatitis virus C glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. 90: 8033–8037. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- F32 AI10523/AI/NIAID NIH HHS/United States
- F32 CA93033-01/CA/NCI NIH HHS/United States
- K01 CA094223/CA/NCI NIH HHS/United States
- R01CA78461-04/CA/NCI NIH HHS/United States
- P01 CA042063/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- P30-CA14051/CA/NCI NIH HHS/United States
- R37-GM34277/GM/NIGMS NIH HHS/United States
- R01 CA078461/CA/NCI NIH HHS/United States
- R37 GM034277/GM/NIGMS NIH HHS/United States
- R01-AI32486/AI/NIAID NIH HHS/United States
- F32 AI010523/AI/NIAID NIH HHS/United States
- F32 CA093033/CA/NCI NIH HHS/United States
- KO1CA94223/CA/NCI NIH HHS/United States
- P01-CA42063/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous