Majority of divergence between closely related DNA samples is due to indels - PubMed (original) (raw)
Comparative Study
. 2003 Apr 15;100(8):4661-5.
doi: 10.1073/pnas.0330964100. Epub 2003 Apr 2.
Affiliations
- PMID: 12672966
- PMCID: PMC153612
- DOI: 10.1073/pnas.0330964100
Comparative Study
Majority of divergence between closely related DNA samples is due to indels
Roy J Britten et al. Proc Natl Acad Sci U S A. 2003.
Abstract
It was recently shown that indels are responsible for more than twice as many unmatched nucleotides as are base substitutions between samples of chimpanzee and human DNA. A larger sample has now been examined and the result is similar. The number of indels is approximately 1/12th of the number of base substitutions and the average length of the indels is 36 nt, including indels up to 10 kb. The ratio (R(u)) of unpaired nucleotides attributable to indels to those attributable to substitutions is 3.0 for this 2 million-nt chimp DNA sample compared with human. There is similar evidence of a large value of R(u) for sea urchins from the polymorphism of a sample of Strongylocentrotus purpuratus DNA (R(u) = 3-4). Other work indicates that similarly, per nucleotide affected, large differences are seen for indels in the DNA polymorphism of the plant Arabidopsis thaliana (R(u) = 51). For the insect Drosophila melanogaster a high value of R(u) (4.5) has been determined. For the nematode Caenorhabditis elegans the polymorphism data are incomplete but high values of R(u) are likely. Comparison of two strains of Escherichia coli O157:H7 shows a preponderance of indels. Because these six examples are from very distant systematic groups the implication is that in general, for alignments of closely related DNA, indels are responsible for many more unmatched nucleotides than are base substitutions. Human genetic evidence suggests that indels are a major source of gene defects, indicating that indels are a significant source of evolutionary change.
Figures
Figure 1
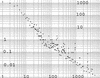
The raw data on gaps between chimp and human alignments. Shown is log–log plot of number of gaps of a given size as a function of size. The vertical axis is the number of gaps and the horizontal axis is the gap length in nucleotides. The line near the bottom is all of the larger gaps, which are present only once with a given length. Gaps >5 kb are uncertain.
Figure 2
The density of gaps vs. gap size. Shown is a log–log plot of the density function D k against gap size. The horizontal axis is gap length in nucleotides. The vertical axis is the density function, which is the number of gaps of a given size divided by the spacing in length between gaps, which is the average of the difference in length to the next smaller gap and the difference in length to the next larger gap. Shown are gaps <5 kb.
Figure 3
The cumulative total of the length of gaps vs. gap size. The number of gaps of a given size is multiplied by the length of the gap and added to the previous total to obtain the cumulative total. The horizontal logarithmic axis is the gap size and the vertical logarithmic axis is the cumulative total. It is clear that the larger gaps contribute heavily. The last four points represent sparse data because long gaps are difficult to measure. New data could easily raise this part of the curve.
Similar articles
- Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels.
Britten RJ. Britten RJ. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13633-5. doi: 10.1073/pnas.172510699. Epub 2002 Oct 4. Proc Natl Acad Sci U S A. 2002. PMID: 12368483 Free PMC article. - An evolutionary constraint: strongly disfavored class of change in DNA sequence during divergence of cis-regulatory modules.
Cameron RA, Chow SH, Berney K, Chiu TY, Yuan QA, Krämer A, Helguero A, Ransick A, Yun M, Davidson EH. Cameron RA, et al. Proc Natl Acad Sci U S A. 2005 Aug 16;102(33):11769-74. doi: 10.1073/pnas.0505291102. Epub 2005 Aug 8. Proc Natl Acad Sci U S A. 2005. PMID: 16087870 Free PMC article. - Comparative genomic analysis of human and chimpanzee indicates a key role for indels in primate evolution.
Wetterbom A, Sevov M, Cavelier L, Bergström TF. Wetterbom A, et al. J Mol Evol. 2006 Nov;63(5):682-90. doi: 10.1007/s00239-006-0045-7. Epub 2006 Oct 29. J Mol Evol. 2006. PMID: 17075697 - Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli.
Reid SD, Selander RK, Whittam TS. Reid SD, et al. J Bacteriol. 1999 Jan;181(1):153-60. doi: 10.1128/JB.181.1.153-160.1999. J Bacteriol. 1999. PMID: 9864325 Free PMC article. - [Chimpanzee genome sequencing and comparative analysis with the human genome].
Watanabe H, Hattori M. Watanabe H, et al. Tanpakushitsu Kakusan Koso. 2006 Feb;51(2):178-87. Tanpakushitsu Kakusan Koso. 2006. PMID: 16457209 Review. Japanese. No abstract available.
Cited by
- General continuous-time Markov model of sequence evolution via insertions/deletions: are alignment probabilities factorable?
Ezawa K. Ezawa K. BMC Bioinformatics. 2016 Aug 11;17:304. doi: 10.1186/s12859-016-1105-7. BMC Bioinformatics. 2016. PMID: 27638547 Free PMC article. - Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies.
García-Lor A, Luro F, Navarro L, Ollitrault P. García-Lor A, et al. Mol Genet Genomics. 2012 Jan;287(1):77-94. doi: 10.1007/s00438-011-0658-4. Epub 2011 Dec 11. Mol Genet Genomics. 2012. PMID: 22160318 - 2S albumin g13 polypeptide, less related to Fag e 2, can be eliminated in common buckwheat (Fagopyrum esculentum Moench) seeds.
Monshi FI, Katsube-Tanaka T. Monshi FI, et al. Food Chem (Oxf). 2022 Sep 26;5:100138. doi: 10.1016/j.fochms.2022.100138. eCollection 2022 Dec 30. Food Chem (Oxf). 2022. PMID: 36187231 Free PMC article. - Inter- and intralocus recombination drive MHC class IIB gene diversification in a teleost, the three-spined stickleback Gasterosteus aculeatus.
Reusch TB, Langefors A. Reusch TB, et al. J Mol Evol. 2005 Oct;61(4):531-41. doi: 10.1007/s00239-004-0340-0. Epub 2005 Aug 24. J Mol Evol. 2005. PMID: 16132469 - DNA indels in coding regions reveal selective constraints on protein evolution in the human lineage.
de la Chaux N, Messer PW, Arndt PF. de la Chaux N, et al. BMC Evol Biol. 2007 Oct 12;7:191. doi: 10.1186/1471-2148-7-191. BMC Evol Biol. 2007. PMID: 17935613 Free PMC article.
References
- Gu X, Li W H. J Mol Evol. 1995;40:464–473. - PubMed
- Nickerson E, Nelson D L. Genomics. 1998;50:368–372. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous